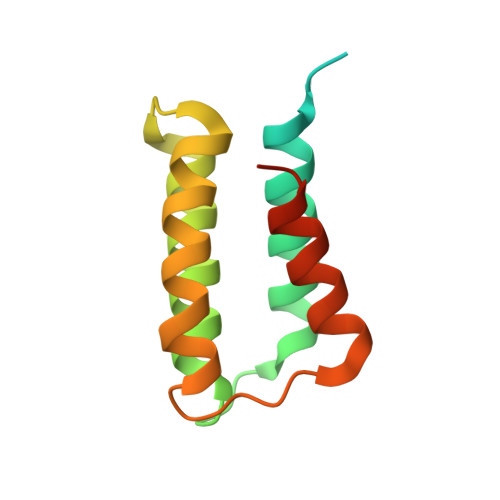
Crystal structure and mutagenic analysis of a bacteriocin immunity protein, Mun-im
Jeon, H.J., Noda, M., Matoba, Y., Kumagai, T., Sugiyama, M.(2009) Biochem Biophys Res Commun 378: 574-578
- PubMed: 19061861
- DOI: https://doi.org/10.1016/j.bbrc.2008.11.093
- Primary Citation of Related Structures:
2ZRR - PubMed Abstract:
Bacteriocin-producing lactic acid bacteria (LAB) possess a self-protection factor, which is generally called an immunity protein. In this study, we determine the crystal structure of an immunity protein, designated Mun-im, which was classified into subgroup B immunity proteins for class IIa bacteriocins. The Mun-im protein takes a left-turning antiparallel four-helix bundle structure with the flexible N- and C-terminal parts. Although the amino acid sequences of the subgroup B immunity proteins are distinguished from those of the subgroup A, the core structure of Mun-im is well-superimposed with that of the subgroup A immunity protein, EntA-im, and the C-terminus of both proteins is flexible. However, the C-terminus of Mun-im is obviously shorter than that of the subgroup A. We found through mutagenic study of Mun-im that the C-terminus and the K86 residue on the helix 4 in the immunity protein molecule are important for expression of the immunity activity on the subgroup B immunity proteins.
- Department of Molecular Microbiology and Biotechnology, Graduate School of Biomedical Sciences, Hiroshima University, Kasumi 1-2-3, Minami-ku, Hiroshima 734-8551, Japan.
Organizational Affiliation: