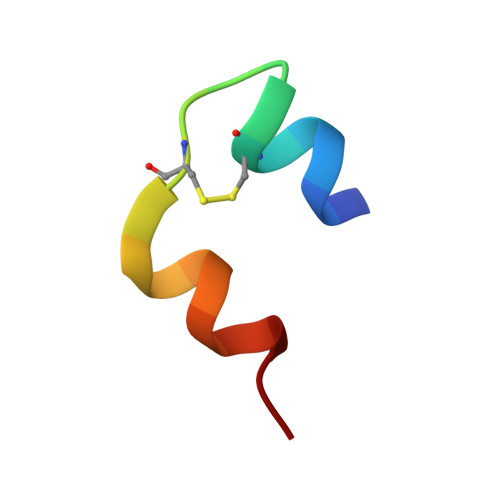
An abnormal pK(a) value of internal histidine of the insulin molecule revealed by neutron crystallographic analysis
Ishikawa, T., Chatake, T., Morimoto, Y., Maeda, M., Kurihara, K., Tanaka, I., Niimura, N.(2008) Biochem Biophys Res Commun 376: 32-35
- PubMed: 18725203
- DOI: https://doi.org/10.1016/j.bbrc.2008.08.071
- Primary Citation of Related Structures:
2ZPP - PubMed Abstract:
Insulin is stored in pancreatic beta-cell as hexameric form with Zn2+ ions, while the hormonally active form is monomer. The hexamer requires the coordination of Zn2+ ions to the HisB10. In order to reveal the mechanism of the hexamerization of insulin, we investigated the Zn2+ free insulin at pD6.6 and pD9 by neutron crystallographic analyses. HisB10 is doubly protonated not only at pD6.6 but also at pD9, indicating an abnormal pK(a) of this histidine. It is suggested that HisB10 acts on a strong cation capture and contributes to the high stability of the hexameric form in pancreas.
- Kyoto University Research Reactor, Asashironishi 2, Kumatori, Osaka 590-0494, Japan.
Organizational Affiliation: