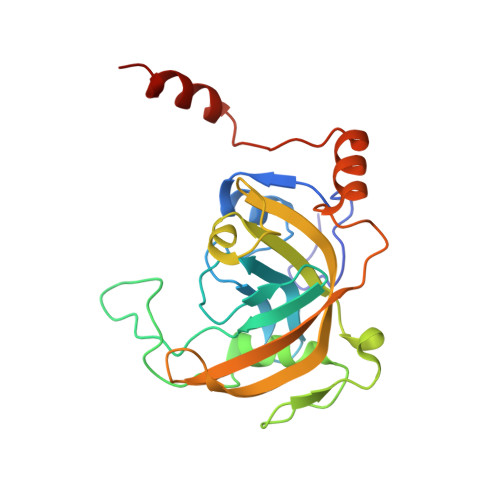
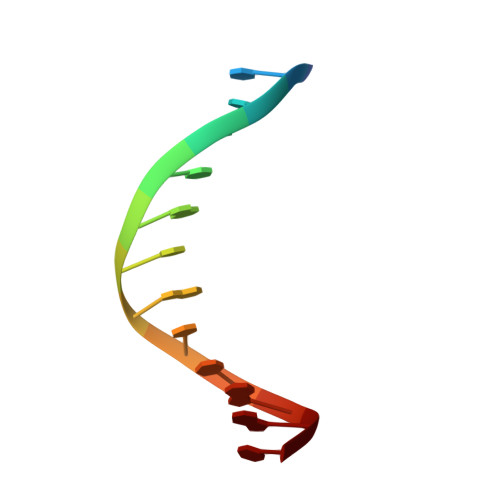
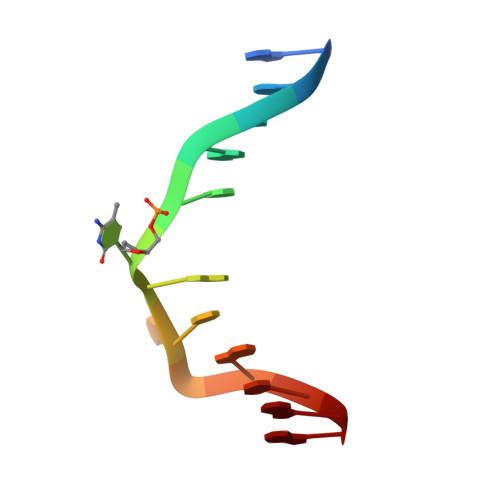
The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Hashimoto, H., Horton, J.R., Zhang, X., Bostick, M., Jacobsen, S.E., Cheng, X.(2008) Nature 455: 826-829
- PubMed: 18772888
- DOI: https://doi.org/10.1038/nature07280
- Primary Citation of Related Structures:
2ZO0, 2ZO1, 2ZO2 - PubMed Abstract:
Maintenance methylation of hemimethylated CpG dinucleotides at DNA replication forks is the key to faithful mitotic inheritance of genomic methylation patterns. UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) is required for maintenance methylation by interacting with DNA nucleotide methyltransferase 1 (DNMT1), the maintenance methyltransferase, and with hemimethylated CpG, the substrate for DNMT1 (refs 1 and 2). Here we present the crystal structure of the SET and RING-associated (SRA) domain of mouse UHRF1 in complex with DNA containing a hemimethylated CpG site. The DNA is contacted in both the major and minor grooves by two loops that penetrate into the middle of the DNA helix. The 5-methylcytosine has flipped completely out of the DNA helix and is positioned in a binding pocket with planar stacking contacts, Watson-Crick polar hydrogen bonds and van der Waals interactions specific for 5-methylcytosine. Hence, UHRF1 contains a previously unknown DNA-binding module and is the first example of a non-enzymatic, sequence-specific DNA-binding protein domain to use the base flipping mechanism to interact with DNA.
- Department of Biochemistry, Emory University School of Medicine, 1510 Clifton Road, Atlanta, Georgia 30322, USA.
Organizational Affiliation: