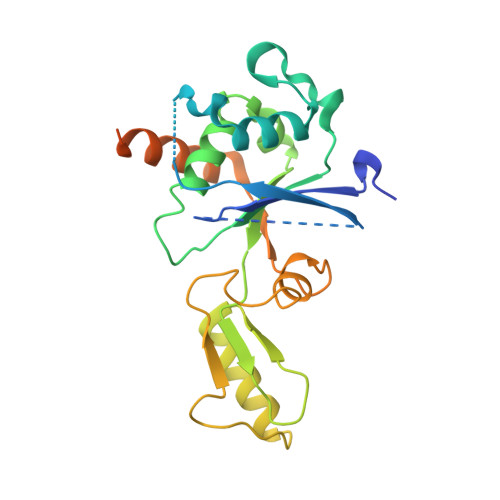
Crystal structure of DNA gyrase B' domain sheds lights on the mechanism for T-segment navigation
Fu, G.S., Wu, J.J., Liu, W., Zhu, D.Y., Hu, Y.L., Deng, J., Zhang, X.E., Bi, L.J., Wang, D.C.(2009) Nucleic Acids Res 37: 5908-5916
- PubMed: 19596812
- DOI: https://doi.org/10.1093/nar/gkp586
- Primary Citation of Related Structures:
2ZJT - PubMed Abstract:
DNA gyrase is an indispensible marvelous molecular machine in manipulating the DNA topology for the prokaryotes. In the 'two-gate' mechanism of DNA topoisomerase, T-segment navigation from N- to DNA-gate is a critical step, but the structural basis supporting this scheme is unclear. The crystal structure of DNA gyrase B' subfragment from Mycobacterium tuberculosis reveals an intrinsic homodimer. The two subunits, each consisting of a Tail and a Toprim domain, are tightly packed one another to form a 'crab-like' organization never observed previously from yeast topo II. Structural comparisons show two orientational alterations of the Tail domain, which may be dominated by a 43-residue peptide at the B' module C-terminus. A highly conserved pentapeptide mediates large-scale intrasubunit conformational change as a hinge point. Mutational studies highlight the significant roles of a negatively charge cluster on a groove at dimer interface. On the basis of structural analysis and mutation experiments, a sluice-like model for T-segment transport is proposed.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.
Organizational Affiliation: