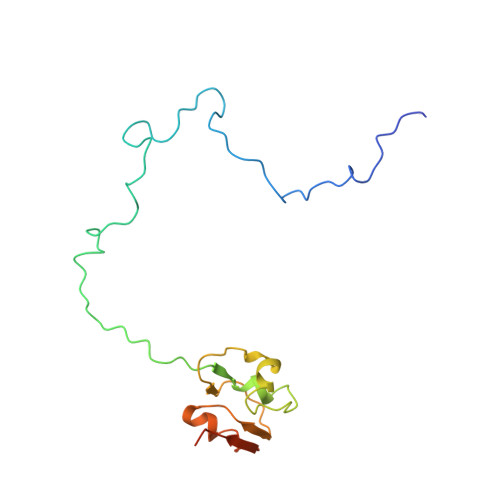
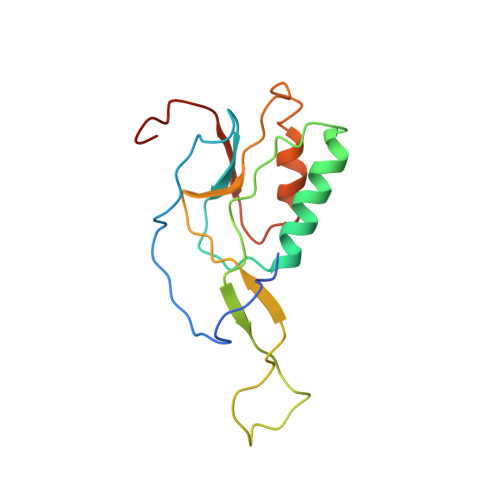
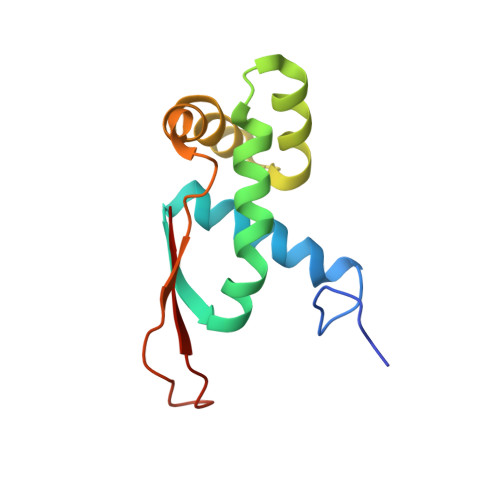
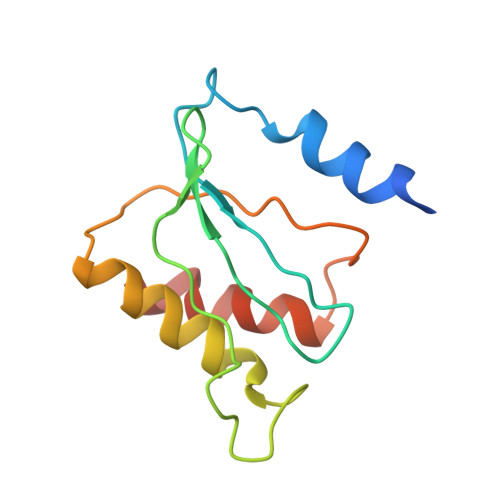
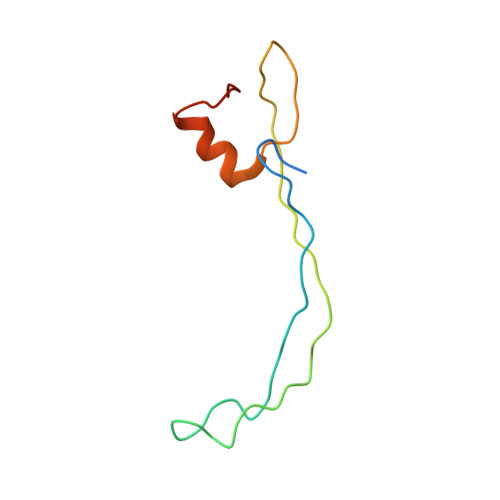
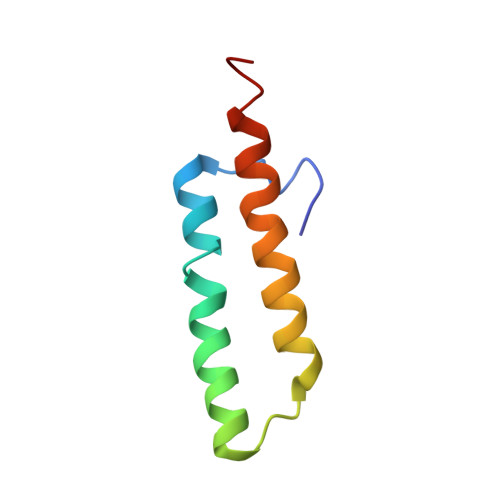
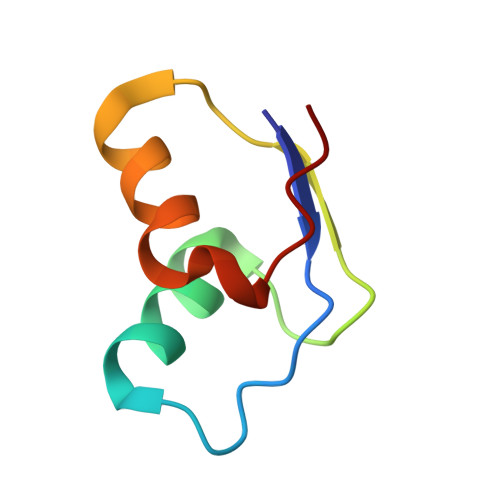
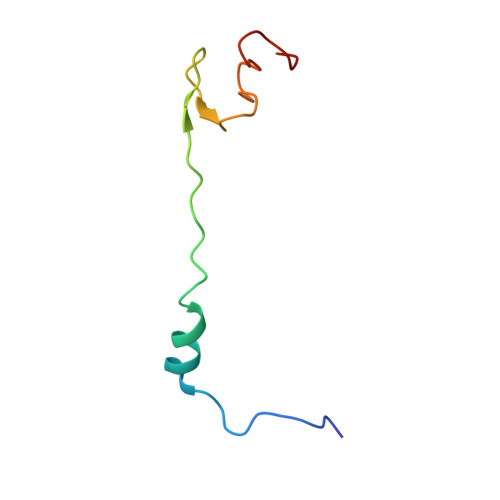
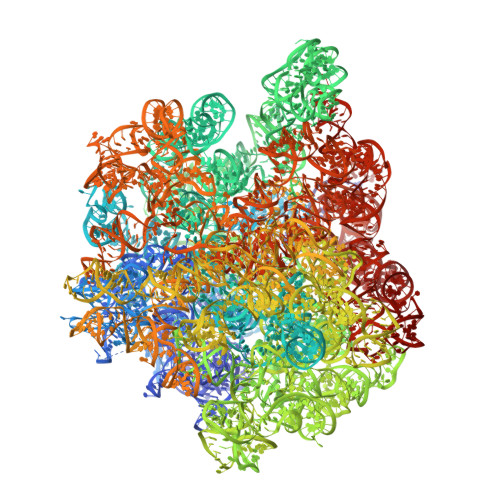
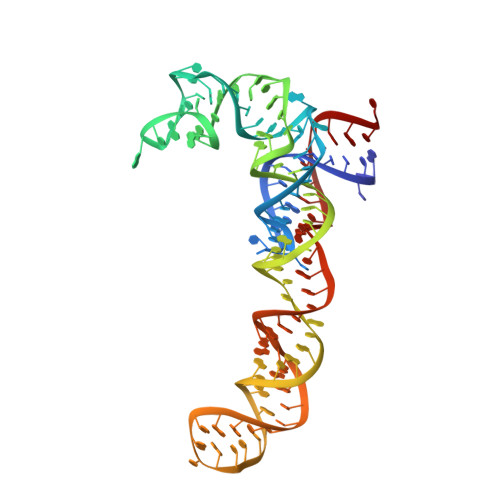
Translational Regulation Via L11: Molecular Switches on the Ribosome Turned on and Off by Thiostrepton and Micrococcin.
Harms, J.M., Wilson, D.N., Schluenzen, F., Connell, S.R., Stachelhaus, T., Zaborowska, Z., Spahn, C.M., Fucini, P.(2008) Mol Cell 30: 26
- PubMed: 18406324
- DOI: https://doi.org/10.1016/j.molcel.2008.01.009
- Primary Citation of Related Structures:
2ZJP, 2ZJQ, 2ZJR, 3CF5 - PubMed Abstract:
The thiopeptide class of antibiotics targets the GTPase-associated center (GAC) of the ribosome to inhibit translation factor function. Using X-ray crystallography, we have determined the binding sites of thiostrepton (Thio), nosiheptide (Nosi), and micrococcin (Micro), on the Deinococcus radiodurans large ribosomal subunit. The thiopeptides, by binding within a cleft located between the ribosomal protein L11 and helices 43 and 44 of the 23S rRNA, overlap with the position of domain V of EF-G, thus explaining how this class of drugs perturbs translation factor binding to the ribosome. The presence of Micro leads to additional density for the C-terminal domain (CTD) of L7, adjacent to and interacting with L11. The results suggest that L11 acts as a molecular switch to control L7 binding and plays a pivotal role in positioning one L7-CTD monomer on the G' subdomain of EF-G to regulate EF-G turnover during protein synthesis.
- Cluster of Excellence for Macromolecular Complexes, Institut für Organische Chemie und Chemische Biologie, J.W. Goethe-Universität Frankfurt am Main, Max-von-Laue-Strasse 7, D-60438 Frankfurt am Main, Germany.
Organizational Affiliation: