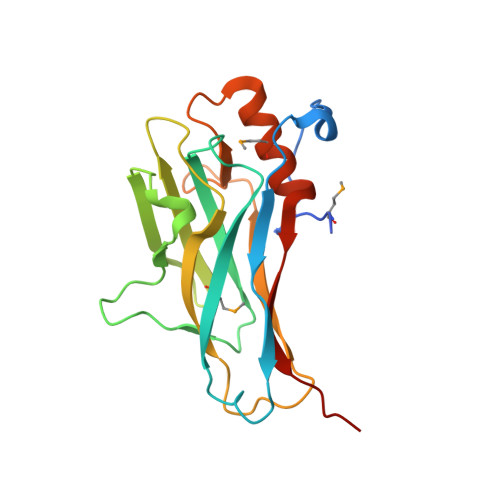
Cohesin diversity revealed by the crystal structure of the anchoring cohesin from Ruminococcus flavefaciens.
Alber, O., Noach, I., Rincon, M.T., Flint, H.J., Shimon, L.J.W., Lamed, R., Frolow, F., Bayer, E.A.(2009) Proteins 77: 699-709
- PubMed: 19544570
- DOI: https://doi.org/10.1002/prot.22483
- Primary Citation of Related Structures:
2ZF9 - PubMed Abstract:
The cellulosome is an intriguing multienzyme complex found in cellulolytic bacteria that plays a key role in the degradation of plant cell-wall polysaccharides. In Ruminococcus flavefaciens, a predominant fiber-degrading bacterium found in ruminants, the cellulosome is anchored to the bacterial cell wall through a relatively short ScaE scaffoldin. Determination of the crystal structure of the lone type-III ScaE cohesin from R. flavefaciens (Rf-CohE) was initiated as a part of a structural effort to define cellulosome assembly. The structure was determined at 1.95 A resolution by single-wavelength anomalous diffraction. This is the first detailed description of a crystal structure for a type-III cohesin, and its features were compared with those of the known type-I and type-II cohesin structures. The Rf-CohE module folds into a nine-stranded beta-sandwich with jellyroll topology, typically observed for cohesins, and includes two beta-flaps in the midst of beta-strands 4 and 8, similar to the type-II cohesin structures. However, the presence in Rf-CohE of an additional 13-residue alpha-helix located between beta-strands 8 and 9 represents a dramatic divergence from other known cohesin structures. The prominent alpha-helix is enveloped by an extensive N-terminal loop, not observed in any other known cohesin, which embraces the helix presumably enhancing its stability. A planar surface at the upper portion of the front face of the molecule, bordered by beta-flap 8, exhibits plausible dimensions and exposed amino acid residues to accommodate the dockerin-binding site.
- Department of Biological Chemistry, The Weizmann Institute of Science, Rehovot 76100, Israel.
Organizational Affiliation: