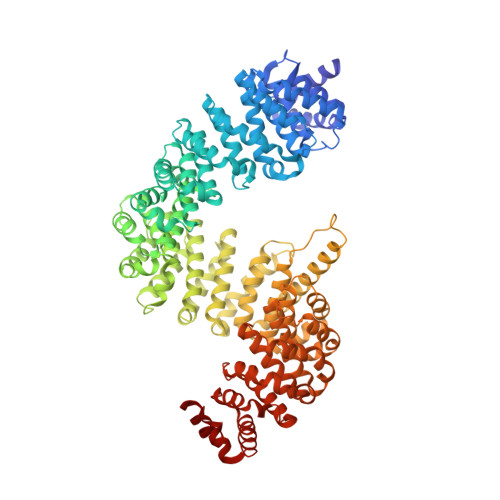
Structural basis for substrate recognition and dissociation by human transportin 1
Imasaki, T., Shimizu, T., Hashimoto, H., Hidaka, Y., Kose, S., Imamoto, N., Yamada, M., Sato, M.(2007) Mol Cell 28: 57-67
- PubMed: 17936704
- DOI: https://doi.org/10.1016/j.molcel.2007.08.006
- Primary Citation of Related Structures:
2Z5J, 2Z5K, 2Z5M, 2Z5N, 2Z5O - PubMed Abstract:
Transportin 1 (Trn1) is a transport receptor that transports substrates from the cytoplasm to the nucleus through nuclear pore complexes by recognizing nuclear localization signals (NLSs). Here we describe four crystal structures of human Trn1 in a substrate-free form as well as in the complex with three NLSs (hnRNP D, JKTBP, and TAP, respectively). Our data have revealed that (1) Trn1 has two sites for binding NLSs, one with high affinity (site A) and one with low affinity (site B), and NLS interaction at site B controls overall binding affinity for Trn1; (2) Trn1 recognizes the NLSs at site A followed by conformational change at site B to interact with the NLSs; and (3) a long flexible loop, characteristic of Trn1, interacts with site B, thereby displacing transport substrate in the nucleus. These studies provide deep understanding of substrate recognition and dissociation by Trn1 in import pathways.
- Field of Supramolecular Biology, International Graduate School of Arts and Sciences, Yokohama City University, 1-7-29 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan.
Organizational Affiliation: