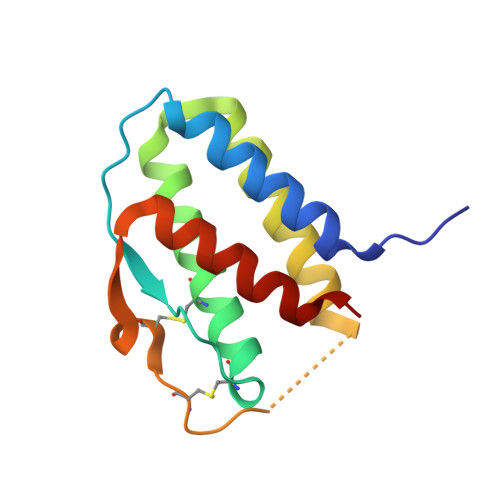
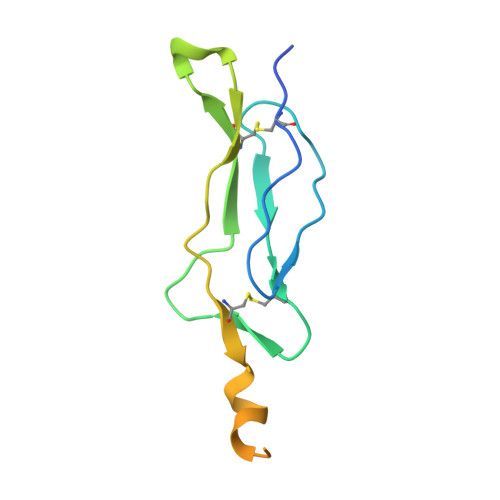
Crystal structure of the IL-15-IL-15Ralpha complex, a cytokine-receptor unit presented in trans
Chirifu, M., Hayashi, C., Nakamura, T., Toma, S., Shuto, T., Kai, H., Yamagata, Y., Davis, S.J., Ikemizu, S.(2007) Nat Immunol 8: 1001-1007
- PubMed: 17643103
- DOI: https://doi.org/10.1038/ni1492
- Primary Citation of Related Structures:
2Z3Q, 2Z3R - PubMed Abstract:
Interleukin 15 (IL-15) and IL-2, which promote the survival of memory CD8(+) T cells and regulatory T cells, respectively, bind receptor complexes that share beta- and gamma-signaling subunits. Receptor specificity is provided by unique, nonsignaling alpha-subunits. Whereas IL-2 receptor-alpha (IL-2Ralpha) is expressed together in cis with the beta- and gamma-subunits on T cells and B cells, IL-15Ralpha is expressed in trans on antigen-presenting cells. Here we present a 1.85-A crystal structure of the human IL-15-IL-15Ralpha complex. The structure provides insight into the molecular basis of the specificity of cytokine recognition and emphasizes the importance of water in generating this very high-affinity complex. Despite very low IL-15-IL-2 sequence homology and distinct receptor architecture, the topologies of the IL-15-IL-15Ralpha and IL-2-IL-2Ralpha complexes are very similar. Our data raise the possibility that IL-2, like IL-15, might be capable of being presented in trans in the context of its unique receptor alpha-chain.
- Graduate School of Pharmaceutical Sciences, Kumamoto University, 5-1 Oe-honmachi, Kumamoto 862-0973, Japan.
Organizational Affiliation: