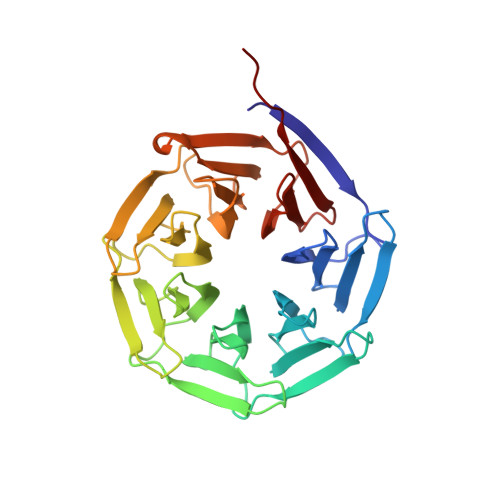
Structural basis for streptogramin B resistance in Staphylococcus aureus by virginiamycin B lyase
Korczynska, M., Mukhtar, T.A., Wright, G.D., Berghuis, A.M.(2007) Proc Natl Acad Sci U S A 104: 10388-10393
- PubMed: 17563376
- DOI: https://doi.org/10.1073/pnas.0701809104
- Primary Citation of Related Structures:
2Z2N, 2Z2O, 2Z2P - PubMed Abstract:
The streptogramin combination therapy of quinupristin-dalfopristin (Synercid) is used to treat infections caused by bacterial pathogens, such as methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus faecium. However, the effectiveness of this therapy is being compromised because of an increased incidence of streptogramin resistance. One of the clinically observed mechanisms of resistance is enzymatic inactivation of the type B streptogramins, such as quinupristin, by a streptogramin B lyase, i.e., virginiamycin B lyase (Vgb). The enzyme catalyzes the linearization of the cyclic antibiotic via a cleavage that requires a divalent metal ion. Here, we present crystal structures of Vgb from S. aureus in its apoenzyme form and in complex with quinupristin and Mg2+ at 1.65- and 2.8-A resolution, respectively. The fold of the enzyme is that of a seven-bladed beta-propeller, although the sequence reveals no similarity to other known members of this structural family. Quinupristin binds to a large depression on the surface of the enzyme, where it predominantly forms van der Waals interactions. Validated by site-directed mutagenesis studies, a reaction mechanism is proposed in which the initial abstraction of a proton is facilitated by a Mg2+ -linked conjugated system. Analysis of the Vgb-quinupristin structure and comparison with the complex between quinupristin and its natural target, the 50S ribosomal subunit, reveals features that can be exploited for developing streptogramins that are impervious to Vgb-mediated resistance.
- Department of Biochemistry, McGill University, Montreal, QC, Canada H3A 4A2.
Organizational Affiliation: