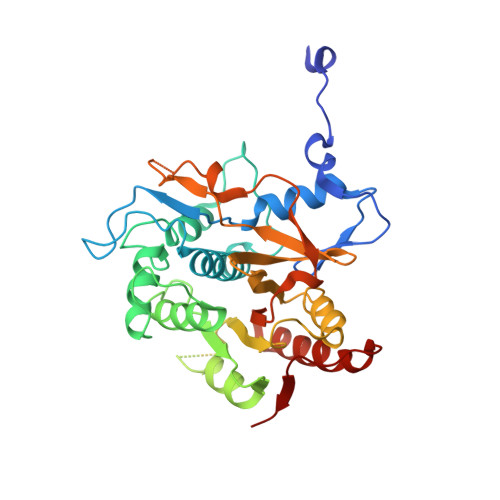
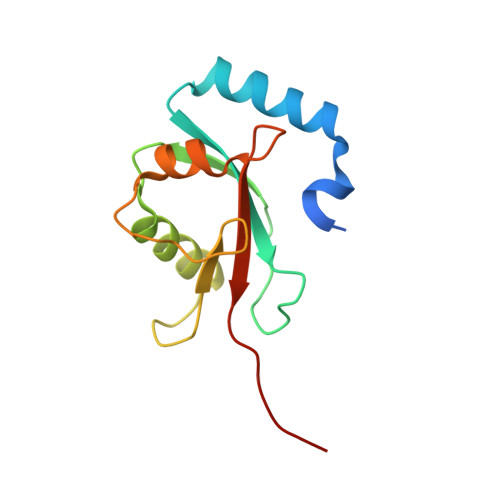
The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy.
Satoo, K., Noda, N.N., Kumeta, H., Fujioka, Y., Mizushima, N., Ohsumi, Y., Inagaki, F.(2009) EMBO J 28: 1341-1350
- PubMed: 19322194
- DOI: https://doi.org/10.1038/emboj.2009.80
- Primary Citation of Related Structures:
2Z0D, 2Z0E, 2ZZP - PubMed Abstract:
Atg8 is conjugated to phosphatidylethanolamine (PE) by ubiquitin-like conjugation reactions. Atg8 has at least two functions in autophagy: membrane biogenesis and target recognition. Regulation of PE conjugation and deconjugation of Atg8 is crucial for these functions in which Atg4 has a critical function by both processing Atg8 precursors and deconjugating Atg8-PE. Here, we report the crystal structures of catalytically inert human Atg4B (HsAtg4B) in complex with processed and unprocessed forms of LC3, a mammalian orthologue of yeast Atg8. On LC3 binding, the regulatory loop and the N-terminal tail of HsAtg4B undergo large conformational changes. The regulatory loop masking the entrance of the active site of free HsAtg4B is lifted by LC3 Phe119, so that a groove is formed along which the LC3 tail enters the active site. At the same time, the N-terminal tail masking the exit of the active site of HsAtg4B in the free form is detached from the enzyme core and a large flat surface is exposed, which might enable the enzyme to access the membrane-bound LC3-PE.
- Department of Structural Biology, Graduate School of Pharmaceutical Sciences, Hokkaido University, Sapporo, Japan.
Organizational Affiliation: