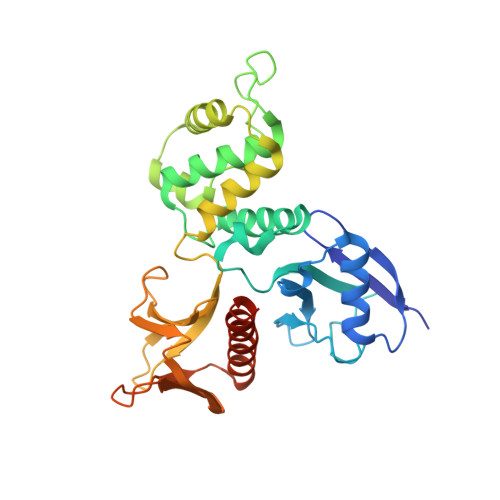
Structural basis for type II membrane protein binding by ERM proteins revealed by the radixin-neutral endopeptidase 24.11 (NEP) complex
Terawaki, S., Kitano, K., Hakoshima, T.(2007) J Biological Chem 282: 19854-19862
- PubMed: 17459884
- DOI: https://doi.org/10.1074/jbc.M609232200
- Primary Citation of Related Structures:
2YVC - PubMed Abstract:
ERM (Ezrin/Radixin/Moesin) proteins mediate formation of membrane-associated cytoskeletons by simultaneously binding actin filaments and the C-terminal cytoplasmic tails of adhesion molecules (type I membrane proteins). ERM proteins also bind neutral endopeptidase 24.11 (NEP), a type II membrane protein, even though the N-terminal cytoplasmic tail of NEP possesses the opposite peptide polarity to that of type I membrane proteins. Here, we determined the crystal structure of the radixin FERM (Four point one and ERM) domain complexed with the N-terminal NEP cytoplasmic peptide. In the FERM-NEP complex, the amphipathic region of the peptide forms a beta strand followed by a hairpin that bind to a shallow groove of FERM subdomain C. NEP binding is stabilized by beta-beta interactions and docking of the NEP hairpin into the hydrophobic pocket of subdomain C. Whereas the binding site of NEP on the FERM domain overlaps with the binding site of intercellular adhesion molecule (ICAM)-2, NEP lacks the Motif-1 sequence conserved in ICAM-2 and related adhesion molecules. The NEP hairpin, although lacking the typical inter-chain hydrogen bond but is stabilized by hydrogen bonds with the main chain and side chains of subdomain C, directs the C-terminal basic region of the NEP peptide away from the groove and toward the membrane. The overlap of the binding sites on subdomain C for NEP and Motif-1 adhesion molecules such as CD44 provides the structural basis for the suppression of cell adhesion through interaction between NEP and ERM proteins.
- Structural Biology Laboratory, Nara Institute of Science and Technology, and CREST, Japan Science and Technology Agency, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan.
Organizational Affiliation: