Solution structure of RNA binding domain in Pre-mRNA-splicing factor RBM22
Kasahara, N., Tsuda, K., Muto, Y., Inoue, M., Kigawa, T., Terada, T., Shirouzu, M., Yokoyama, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Pre-mRNA-splicing factor RBM22 | 85 | Homo sapiens | Mutation(s): 0 Gene Names: RBM22 | 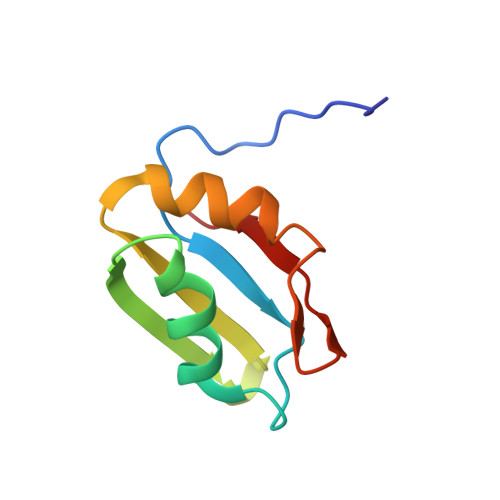 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9NW64 (Homo sapiens) Explore Q9NW64 Go to UniProtKB: Q9NW64 | |||||
PHAROS: Q9NW64 GTEx: ENSG00000086589 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9NW64 | ||||
Sequence AnnotationsExpand | |||||
| |||||