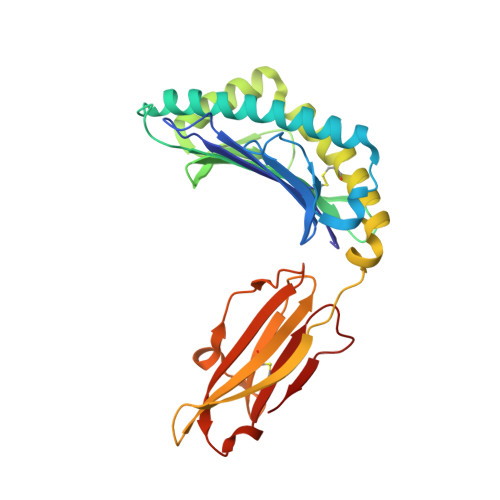
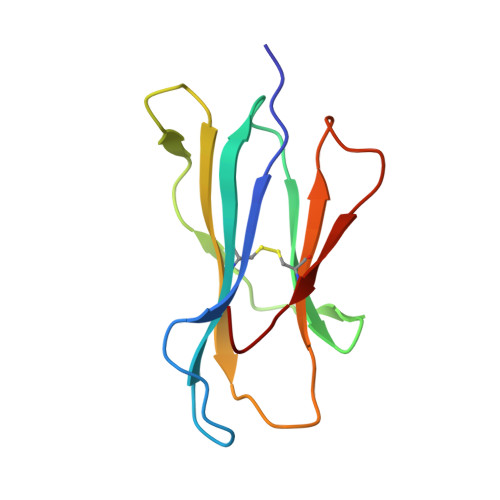
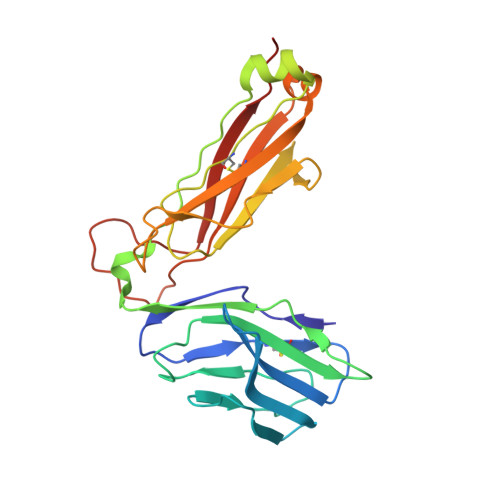
Structural Features Underlying T-Cell Receptor Sensitivity to Concealed Mhc Class I Micropolymorphisms.
Stewart-Jones, G.B., Simpson, P., Anton Van Der Merwe, P., Easterbrook, P., Mcmichael, A.J., Rowland-Jones, S.L., Jones, E.Y., Gillespie, G.M.(2012) Proc Natl Acad Sci U S A 109: E3483
- PubMed: 23161907
- DOI: https://doi.org/10.1073/pnas.1207896109
- Primary Citation of Related Structures:
2YPK, 2YPL - PubMed Abstract:
Polymorphic differences distinguishing MHC class I subtypes often permit the presentation of shared epitopes in conformationally identical formats but can affect T-cell repertoire selection, differentially impacting autoimmune susceptibilities and viral clearance in vivo. The molecular mechanisms underlying this effect are not well understood. We performed structural, thermodynamic, and functional analyses of a conserved T-cell receptor (TCR) which is frequently expanded in response to a HIV-1 epitope when presented by HLA-B*5701 but is not selected by HLA-B*5703, which differs from HLA-B*5701 by two concealed polymorphisms. Our findings illustrate that although both HLA-B*57 subtypes display the epitope in structurally conserved formats, the impact of their polymorphic differences occurs directly as a consequence of TCR ligation, primarily because of peptide adjustments required for TCR binding, which involves the interplay of polymorphic residues and water molecules. These minor differences culminate in subtype-specific differential TCR-binding kinetics and cellular function. Our data demonstrate a potential mechanism whereby the most subtle MHC class I micropolymorphisms can influence TCR use and highlight their implications for disease outcomes.
- Medical Research Council Human Immunology Unit, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, University of Oxford, Oxford OX3 9DS, United Kingdom.
Organizational Affiliation: