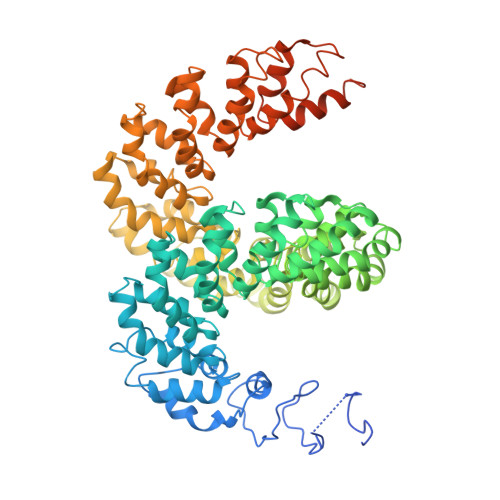
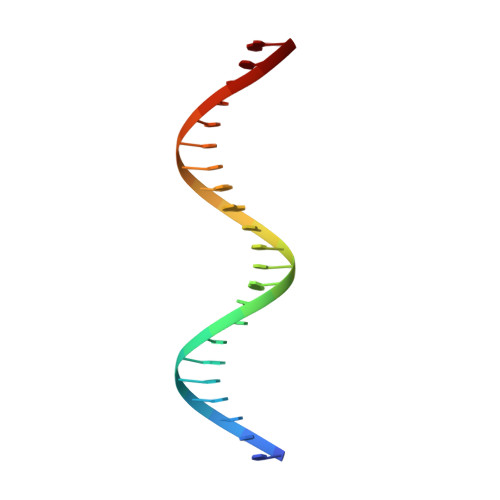
Structure of the Avrbs3-DNA Complex Provides New Insights Into the Initial Thymine-Recognition Mechanism
Stella, S., Molina, R., Yefimenko, I., Prieto, J., Silva, G.H., Bertonati, C., Juillerat, A., Duchateau, P., Montoya, G.(2013) Acta Crystallogr D Biol Crystallogr 69: 1707
- PubMed: 23999294
- DOI: https://doi.org/10.1107/S0907444913016429
- Primary Citation of Related Structures:
2YPF - PubMed Abstract:
Transcription activator-like effectors contain a DNA-binding domain organized in tandem repeats. The repeats include two adjacent residues known as the repeat variable di-residue, which recognize a single base pair, establishing a direct code between the dipeptides and the target DNA. This feature suggests this scaffold as an excellent candidate to generate new protein-DNA specificities for biotechnological applications. Here, the crystal structure of AvrBs3 (residues 152-895, molecular mass 82 kDa) in complex with its target DNA sequence is presented, revealing a new mode of interaction with the initial thymine of the target sequence, together with an analysis of both the binding specificity and the thermodynamic properties of AvrBs3. This study quantifies the affinity and the specificity between AvrBs3 and its target DNA. Moreover, in vitro and in vivo analyses reveal that AvrBs3 does not show a strict nucleotide-binding preference for the nucleotide at the zero position of the DNA, widening the number of possible sequences that could be targeted by this scaffold.
- Structural Biology and Biocomputing Programme, Spanish Cancer Research Centre (CNIO), Melchor Fernandez Almagro, 28029 Madrid, Spain.
Organizational Affiliation: