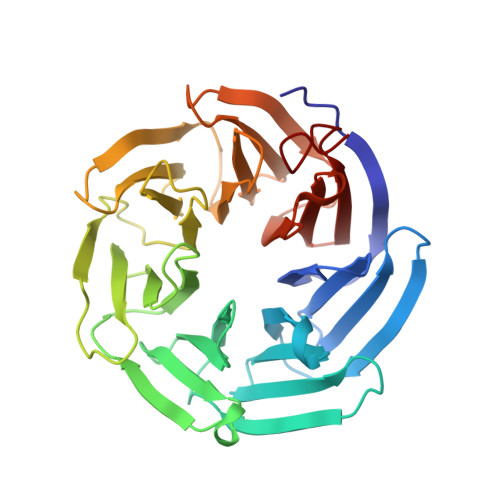
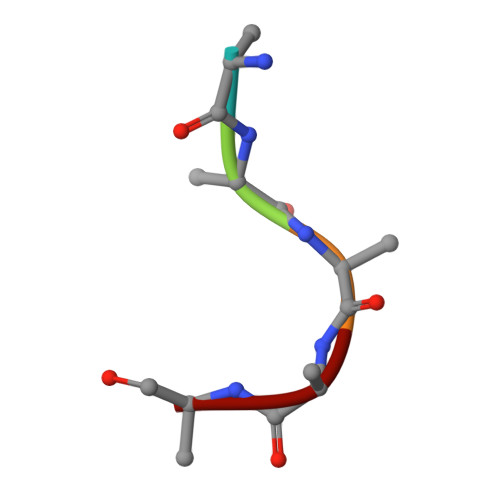
Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Jackson, L.P., Lewis, M., Kent, H.M., Edeling, M.A., Evans, P.R., Duden, R., Owen, D.J.(2012) Dev Cell 23: 1255
- PubMed: 23177648
- DOI: https://doi.org/10.1016/j.devcel.2012.10.017
- Primary Citation of Related Structures:
2YNN, 2YNO, 2YNP - PubMed Abstract:
COPI mediates retrograde trafficking from the Golgi to the endoplasmic reticulum (ER) and within the Golgi stack, sorting transmembrane proteins bearing C-terminal KKxx or KxKxx motifs. The structure of KxKxx motifs bound to the N-terminal WD-repeat domain of β'-COP identifies electrostatic contacts between the motif and complementary patches at the center of the β'-COP propeller. An absolute requirement of a two-residue spacing between the terminal carboxylate group and first lysine residue results from interactions of carbonyl groups in the motif backbone with basic side chains of β'-COP. Similar interactions are proposed to mediate binding of KKxx motifs by the homologous α-COP domain. Mutation of key interacting residues in either domain or in their cognate motifs abolishes in vitro binding and results in mistrafficking of dilysine-containing cargo in yeast without compromising cell viability. Flexibility between β'-COP WD-repeat domains and the location of cargo binding have implications for COPI coat assembly.
- Cambridge Institute for Medical Research, Department of Clinical Biochemistry, University of Cambridge, Hills Road, Cambridge CB2 0XY, UK. lpj21@cam.ac.uk
Organizational Affiliation: