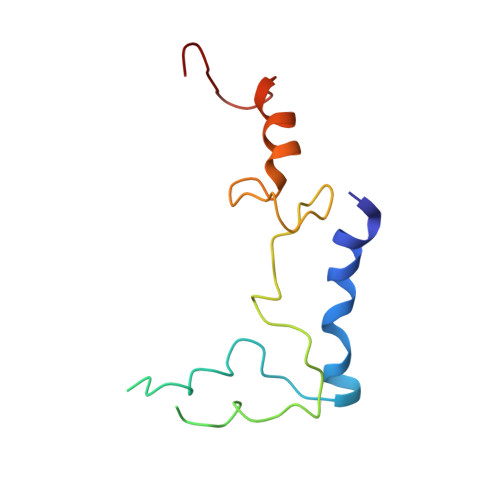
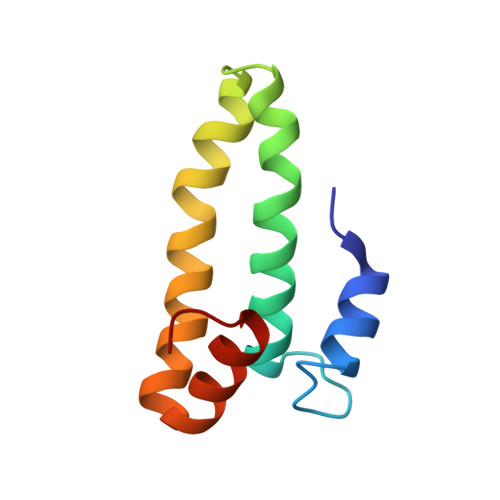
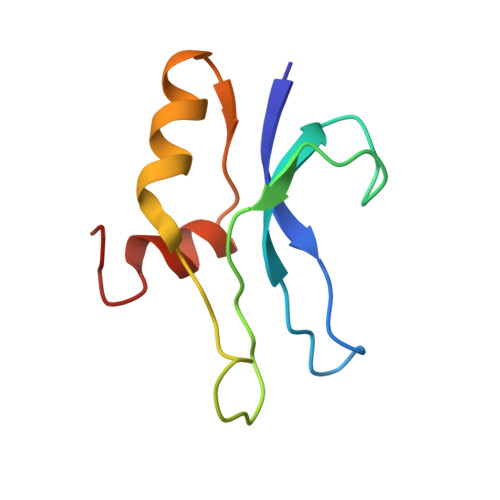
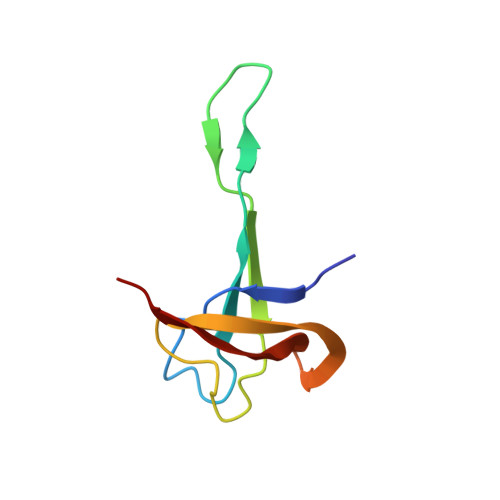
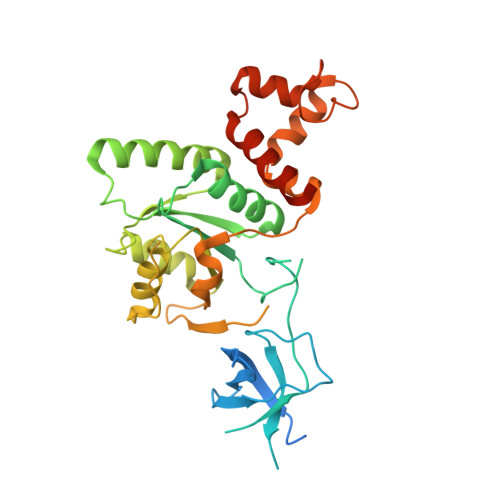
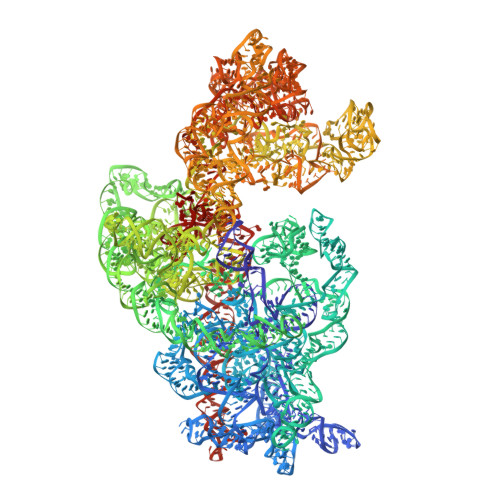
Structural Basis for the Function of a Small Gtpase Rsga on the 30S Ribosomal Subunit Maturation Revealed by Cryoelectron Microscopy.
Guo, Q., Yuan, Y., Xu, Y., Feng, B., Liu, L., Chen, K., Sun, M., Yang, Z., Lei, J., Gao, N.(2011) Proc Natl Acad Sci U S A 108: 13100
- PubMed: 21788480
- DOI: https://doi.org/10.1073/pnas.1104645108
- Primary Citation of Related Structures:
2YKR - PubMed Abstract:
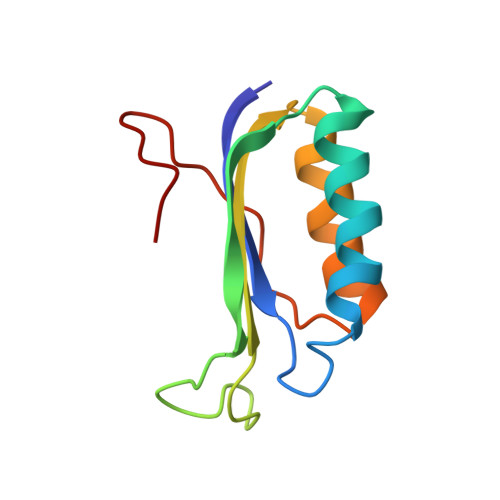
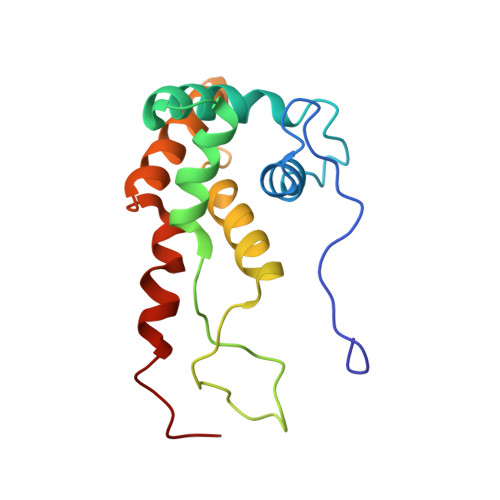
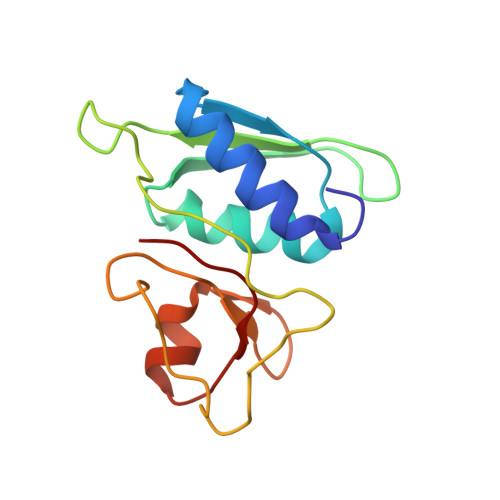
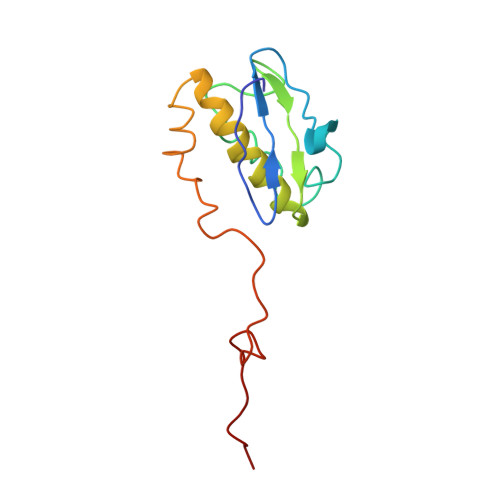
The bacterial RsgA, a circularly permutated GTPase, whose GTPase activity is dependent on the 30S ribosomal subunit, is a late-stage ribosome biogenesis factor involved in the 30S subunit maturation. The role of RsgA is to release another 30S biogenesis factor, RbfA, from the mature 30S subunit in a GTP-dependent manner. Using cryoelectron microscopy, we have determined the structure of the 30S subunit bound with RsgA in the presence of GMPPNP at subnanometer resolution. In the structure, RsgA binds to the central part of the 30S subunit, close to the decoding center, in a position that is incompatible with multiple biogenesis factors, all three translation initiation factors, as well as A-, P-site tRNAs and the 50S subunit. Further structural analysis not only provides a structural model for the RsgA-dependent release of RbfA from the nascent 30S subunit, but also indicates RsgA's role in the ribosomal protein assembly, to promote some tertiary binding protein incorporation. Moreover, together with available biochemical and genetic data, our results suggest that RsgA might be a general checkpoint protein in the late stage of the 30S subunit biogenesis, whose function is not only to release biogenesis factors (e.g., RbfA) from the nascent 30S subunit, but also to block the association of initiation factors to the premature 30S subunit.
- Ministry of Education Protein Science Laboratory, Center for Structural Biology, School of Life Sciences, and Department of Chemistry, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: