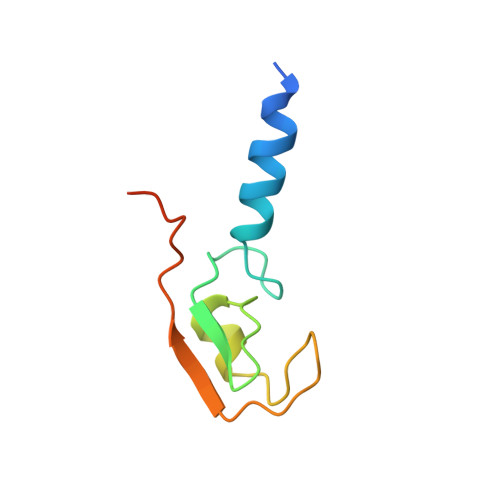
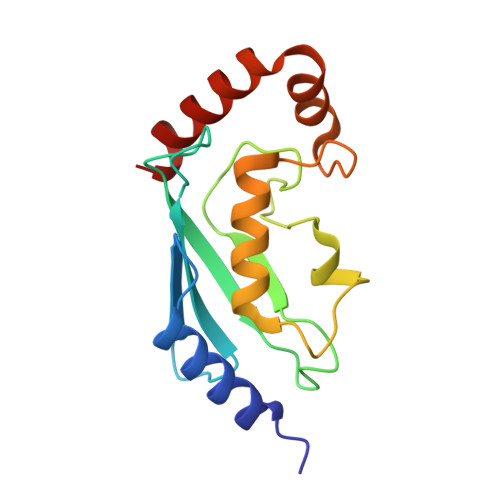
The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Zhang, L., Fairall, L., Goult, B.T., Calkin, A.C., Hong, C., Millard, C.J., Tontonoz, P., Schwabe, J.W.(2011) Genes Dev 25: 1262
- PubMed: 21685362
- DOI: https://doi.org/10.1101/gad.2056211
- Primary Citation of Related Structures:
2YHN, 2YHO - PubMed Abstract:
We previously identified the E3 ubiquitin ligase IDOL as a sterol-dependent regulator of the LDL receptor (LDLR). The molecular pathway underlying IDOL action, however, remains to be determined. Here we report the identification and biochemical and structural characterization of an E2-E3 ubiquitin ligase complex for LDLR degradation. We identified the UBE2D family (UBE2D1-4) as E2 partners for IDOL that support both autoubiquitination and IDOL-dependent ubiquitination of the LDLR in a cell-free system. NMR chemical shift mapping and a 2.1 Å crystal structure of the IDOL RING domain-UBE2D1 complex revealed key interactions between the dimeric IDOL protein and the E2 enzyme. Analysis of the IDOL-UBE2D1 interface also defined the stereochemical basis for the selectivity of IDOL for UBE2Ds over other E2 ligases. Structure-based mutations that inhibit IDOL dimerization or IDOL-UBE2D interaction block IDOL-dependent LDLR ubiquitination and degradation. Furthermore, expression of a dominant-negative UBE2D enzyme inhibits the ability of IDOL to degrade the LDLR in cells. These results identify the IDOL-UBE2D complex as an important determinant of LDLR activity, and provide insight into molecular mechanisms underlying the regulation of cholesterol uptake.
- Howard Hughes Medical Institute, University of California at Los Angeles School of Medicine, USA.
Organizational Affiliation: