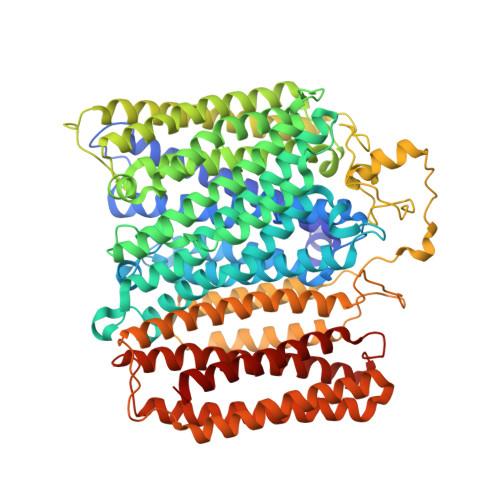
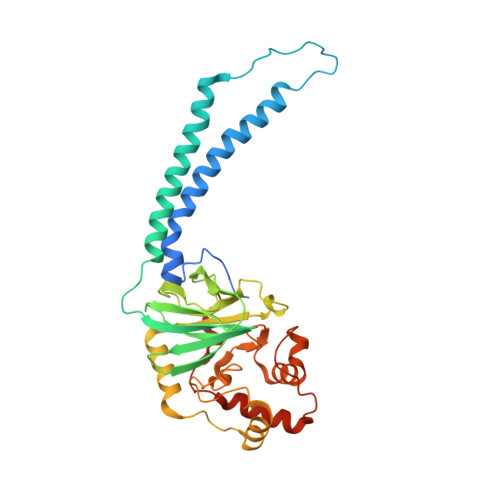
Structural Insights Into Electron Transfer in Caa3-Type Cytochrome Oxidases.
Lyons, J.A., Aragao, D., Slattery, O., Pisliakov, A.V., Soulimane, T., Caffrey, M.(2012) Nature 487: 514
- PubMed: 22763450
- DOI: https://doi.org/10.1038/nature11182
- Primary Citation of Related Structures:
2YEV - PubMed Abstract:
Cytochrome c oxidase is a member of the haem copper oxidase superfamily (HCO). HCOs function as the terminal enzymes in the respiratory chain of mitochondria and aerobic prokaryotes, coupling molecular oxygen reduction to transmembrane proton pumping. Integral to the enzyme's function is the transfer of electrons from cytochrome c to the oxidase via a transient association of the two proteins. Electron entry and exit are proposed to occur from the same site on cytochrome c. Here we report the crystal structure of the caa3-type cytochrome oxidase from Thermus thermophilus, which has a covalently tethered cytochrome c domain. Crystals were grown in a bicontinuous mesophase using a synthetic short-chain monoacylglycerol as the hosting lipid. From the electron density map, at 2.36 Å resolution, a novel integral membrane subunit and a native glycoglycerophospholipid embedded in the complex were identified. Contrary to previous electron transfer mechanisms observed for soluble cytochrome c, the structure reveals the architecture of the electron transfer complex for the fused cupredoxin/cytochrome c domain, which implicates different sites on cytochrome c for electron entry and exit. Support for an alternative to the classical proton gate characteristic of this HCO class is presented.
- Department of Chemical and Environmental Sciences, University of Limerick, Limerick, Ireland.
Organizational Affiliation: