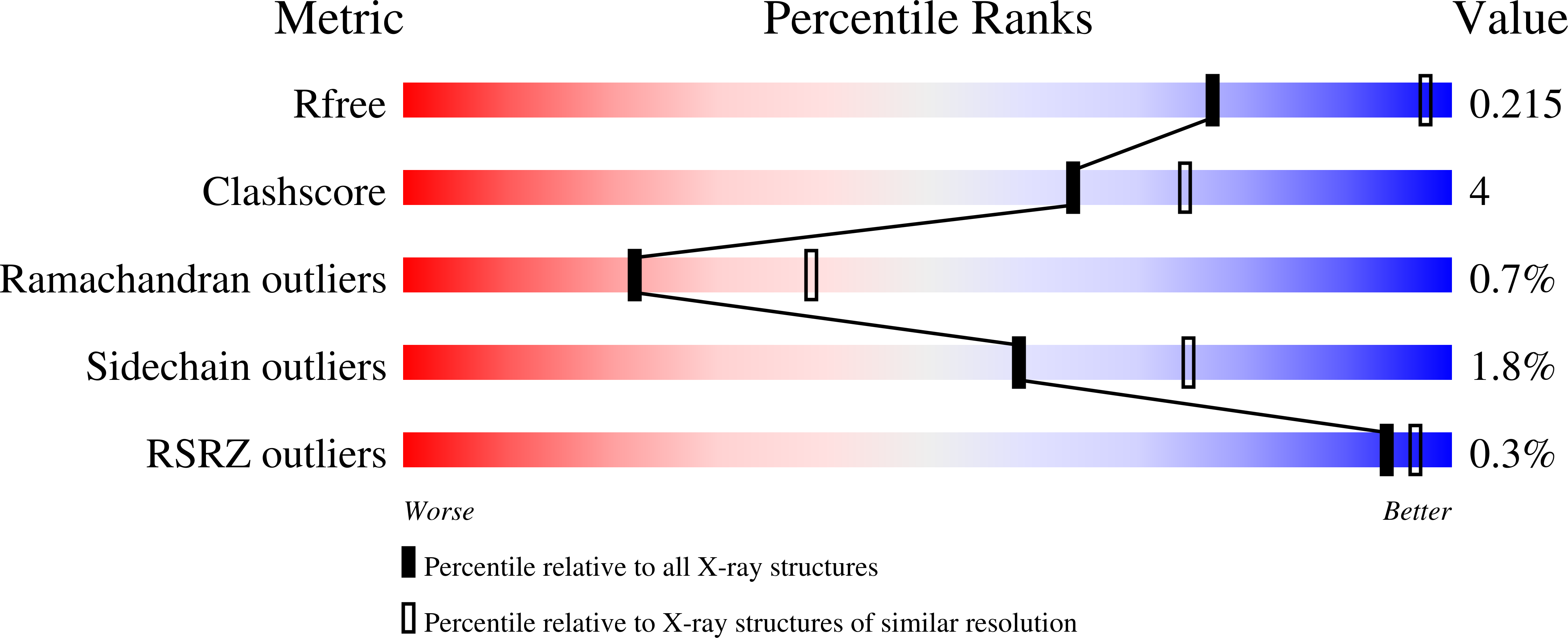
Synergy of Peptide and Sugar in O-Glcnacase Substrate Recognition.
Schimpl, M., Borodkin, V.S., Gray, L.J., Van Aalten, D.M.F.(2012) Chem Biol 19: 173
- PubMed: 22365600
- DOI: https://doi.org/10.1016/j.chembiol.2012.01.011
- Primary Citation of Related Structures:
2YDQ, 2YDR, 2YDS - PubMed Abstract:
Protein O-GlcNAcylation is an essential reversible posttranslational modification in higher eukaryotes. O-GlcNAc addition and removal is catalyzed by O-GlcNAc transferase and O-GlcNAcase, respectively. We report the molecular details of the interaction of a bacterial O-GlcNAcase homolog with three different synthetic glycopeptides derived from characterized O-GlcNAc sites in the human proteome. Strikingly, the peptides bind a conserved O-GlcNAcase substrate binding groove with similar orientation and conformation. In addition to extensive contacts with the sugar, O-GlcNAcase recognizes the peptide backbone through hydrophobic interactions and intramolecular hydrogen bonds, while avoiding interactions with the glycopeptide side chains. These findings elucidate the molecular basis of O-GlcNAcase substrate specificity, explaining how a single enzyme achieves cycling of the complete O-GlcNAc proteome. In addition, this work will aid development of O-GlcNAcase inhibitors that target the peptide binding site.
- Division of Cell Signalling & Immunology, College of Life Sciences, University of Dundee, Dundee DD1 5EH, Scotland.
Organizational Affiliation: