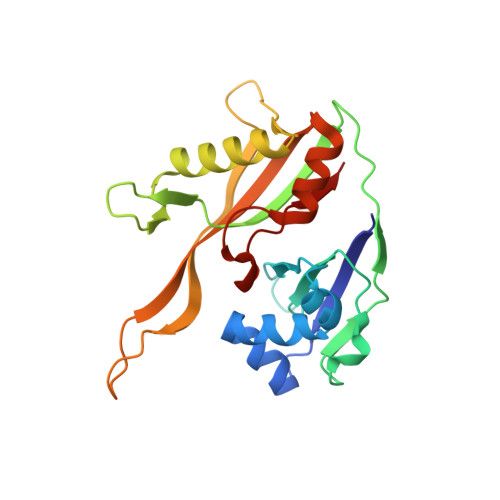
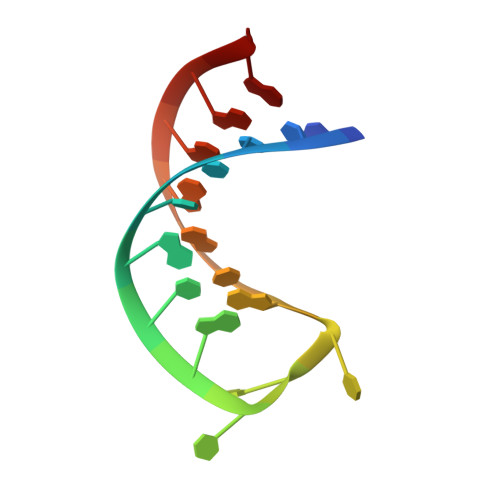
An RNA-Induced Conformational Change Required for Crispr RNA Cleavage by the Endoribonuclease Cse3.
Sashital, D.G., Jinek, M., Doudna, J.A.(2011) Nat Struct Mol Biol 18: 680
- PubMed: 21572442
- DOI: https://doi.org/10.1038/nsmb.2043
- Primary Citation of Related Structures:
2Y8W, 2Y8Y, 2Y9H - PubMed Abstract:
Clustered regularly interspaced short palindromic repeat (CRISPR) chromosomal loci found in prokaryotes provide an adaptive immune system against bacteriophages and plasmids. CRISPR-specific endoRNases produce short RNA molecules (crRNAs) from CRISPR transcripts, which harbor sequences complementary to invasive nucleic acid elements and ensure their selective targeting by CRISPR-associated (Cas) proteins. The extreme sequence divergence of CRISPR-specific endoRNases and their RNA substrates has obscured homology-based comparison of RNA recognition and cleavage mechanisms. Here, we show that Cse3 type CRISPR-specific endoRNases bind a hairpin structure and residues downstream of the cleavage site within the repetitive segment of cognate CRISPR RNA. Cocrystal structures of Cse3-RNA complexes reveal an RNA-induced conformational change in the enzyme active site that aligns the RNA strand for site-specific cleavage. These studies provide insight into a catalytically essential RNA recognition mechanism by a large class of CRISPR-related endoRNases.
- Department of Molecular and Cell Biology, University of California, Berkeley, California, USA.
Organizational Affiliation: