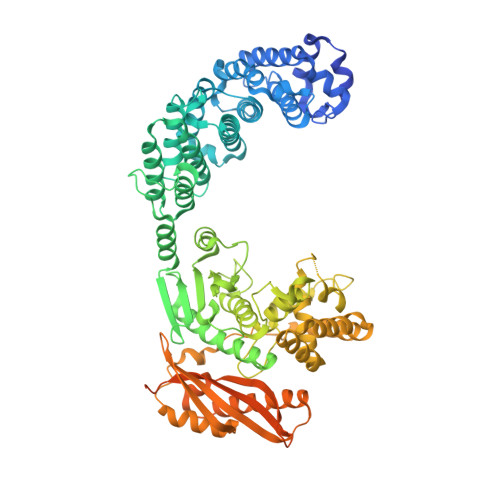
Structure of Collagenase G Reveals a Chew-and -Digest Mechanism of Bacterial Collagenolysis
Eckhard, U., Schoenauer, E., Nuess, D., Brandstetter, H.(2011) Nat Struct Mol Biol 18: 1109
- PubMed: 21947205
- DOI: https://doi.org/10.1038/nsmb.2127
- Primary Citation of Related Structures:
2Y3U, 2Y50, 2Y6I, 2Y72 - PubMed Abstract:
Collagen constitutes one-third of body protein in humans, reflecting its extensive role in health and disease. Of similar importance, therefore, are the idiosyncratic proteases that have evolved for collagen remodeling. The most efficient collagenases are those that enable clostridial bacteria to colonize their host tissues; but despite intense study, the structural and mechanistic basis of these enzymes has remained elusive. Here we present the crystal structure of collagenase G from Clostridium histolyticum at 2.55-Å resolution. By combining the structural data with enzymatic and mutagenesis studies, we derive a conformational two-state model of bacterial collagenolysis, in which recognition and unraveling of collagen microfibrils into triple helices, as well as unwinding of the triple helices, are driven by collagenase opening and closing.
- Division of Structural Biology, Department of Molecular Biology, University of Salzburg, Salzburg, Austria.
Organizational Affiliation: