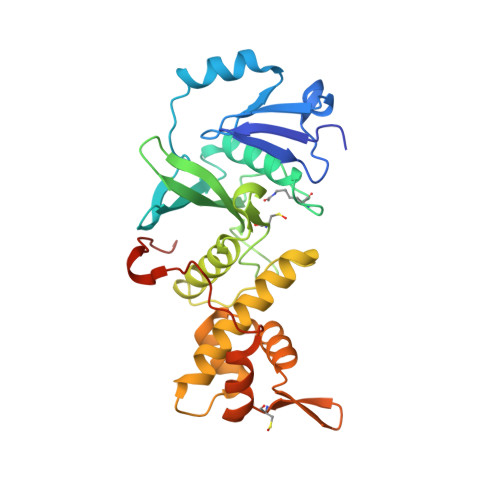
Structural Basis for Mof and Msl3 Recruitment Into the Dosage Compensation Complex by Msl1.
Kadlec, J., Hallacli, E., Lipp, M., Holz, H., Sanchez-Weatherby, J., Cusack, S., Akhtar, A.(2011) Nat Struct Mol Biol 18: 142
- PubMed: 21217699
- DOI: https://doi.org/10.1038/nsmb.1960
- Primary Citation of Related Structures:
2Y0M, 2Y0N - PubMed Abstract:
The male-specific lethal (MSL) complex is required for dosage compensation in Drosophila melanogaster, and analogous complexes exist in mammals. We report structures of binary complexes of mammalian MSL3 and the histone acetyltransferase (HAT) MOF with consecutive segments of MSL1. MSL1 interacts with MSL3 as an extended chain forming an extensive hydrophobic interface, whereas the MSL1-MOF interface involves electrostatic interactions between the HAT domain and a long helix of MSL1. This structure provides insights into the catalytic mechanism of MOF and enables us to show analogous interactions of MOF with NSL1. In Drosophila, selective disruption of Msl1 interactions with Msl3 or Mof severely affects Msl1 targeting to the body of dosage-compensated genes and several high-affinity sites, without affecting promoter binding. We propose that Msl1 acts as a scaffold for MSL complex assembly to achieve specific targeting to the X chromosome.
- European Molecular Biology Laboratory, Grenoble, France.
Organizational Affiliation: