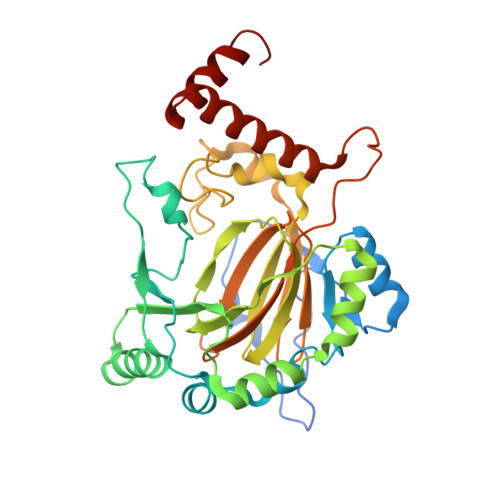
Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains.
Yang, M., Chowdhury, R., Ge, W., Hamed, R.B., McDonough, M.A., Claridge, T.D., Kessler, B.M., Cockman, M.E., Ratcliffe, P.J., Schofield, C.J.(2011) FEBS J 278: 1086-1097
- PubMed: 21251231
- DOI: https://doi.org/10.1111/j.1742-4658.2011.08022.x
- Primary Citation of Related Structures:
2Y0I - PubMed Abstract:
Factor-inhibiting hypoxia-inducible factor (FIH) is an Fe(II)/2-oxoglutarate-dependent dioxygenase that acts as a negative regulator of the hypoxia-inducible factor (HIF) by catalysing β-hydroxylation of an asparaginyl residue in its C-terminal transcriptional activation domain (CAD). In addition to the hypoxia-inducible factor C-terminal transcriptional activation domain (HIF-CAD), FIH also catalyses asparaginyl hydroxylation of many ankyrin repeat domain-containing proteins, revealing a broad sequence selectivity. However, there are few reports on the selectivity of FIH for the hydroxylation of specific residues. Here, we report that histidinyl residues within the ankyrin repeat domain of tankyrase-2 can be hydroxylated by FIH. NMR and crystallographic analyses show that the histidinyl hydroxylation occurs at the β-position. The results further expand the scope of FIH-catalysed hydroxylations.
- Oxford Centre for Integrative Systems Biology, University of Oxford, Oxford, UK.
Organizational Affiliation: