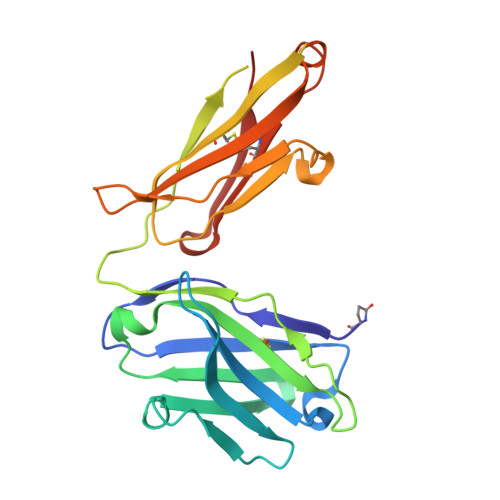
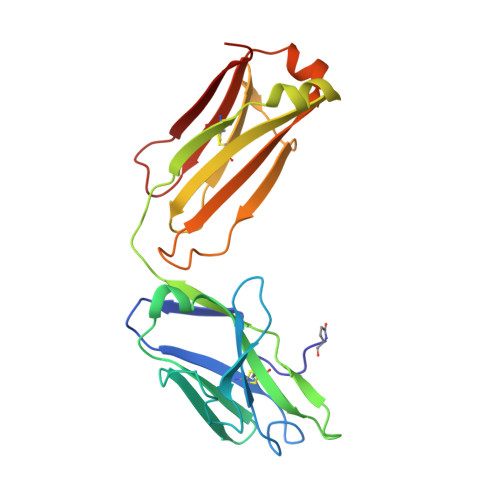
Reactibodies Generated by Kinetic Selection Couple Chemical Reactivity with Favorable Protein Dynamics.
Smirnov, I., Carletti, E., Kurkova, I., Nachon, F., Nicolet, Y., Mitkevich, V.A., Debat, H., Avalle, B., Belogurov, A.A., Kuznetsov, N., Reshetnyak, A., Masson, P., Tonevitsky, A.G., Ponomarenko, N., Makarov, A.A., Friboulet, A., Tramontano, A., Gabibov, A.(2011) Proc Natl Acad Sci U S A 108: 15954
- PubMed: 21896761
- DOI: https://doi.org/10.1073/pnas.1108460108
- Primary Citation of Related Structures:
2XZA, 2XZC - PubMed Abstract:
Igs offer a versatile template for combinatorial and rational design approaches to the de novo creation of catalytically active proteins. We have used a covalent capture selection strategy to identify biocatalysts from within a human semisynthetic antibody variable fragment library that uses a nucleophilic mechanism. Specific phosphonylation at a single tyrosine within the variable light-chain framework was confirmed in a recombinant IgG construct. High-resolution crystallographic structures of unmodified and phosphonylated Fabs display a 15-Å-deep two-chamber cavity at the interface of variable light (V(L)) and variable heavy (V(H)) fragments having a nucleophilic tyrosine at the base of the site. The depth and structure of the pocket are atypical of antibodies in general but can be compared qualitatively with the catalytic site of cholinesterases. A structurally disordered heavy chain complementary determining region 3 loop, constituting a wall of the cleft, is stabilized after covalent modification by hydrogen bonding to the phosphonate tropinol moiety. These features and presteady state kinetics analysis indicate that an induced fit mechanism operates in this reaction. Mutations of residues located in this stabilized loop do not interfere with direct contacts to the organophosphate ligand but can interrogate second shell interactions, because the H3 loop has a conformation adjusted for binding. Kinetic and thermodynamic parameters along with computational docking support the active site model, including plasticity and simple catalytic components. Although relatively uncomplicated, this catalytic machinery displays both stereo- and chemical selectivity. The organophosphate pesticide paraoxon is hydrolyzed by covalent catalysis with rate-limiting dephosphorylation. This reactibody is, therefore, a kinetically selected protein template that has enzyme-like catalytic attributes.
- Institute of Bioorganic Chemistry Russian Academy of Sciences, Moscow 117997, Russia.
Organizational Affiliation: