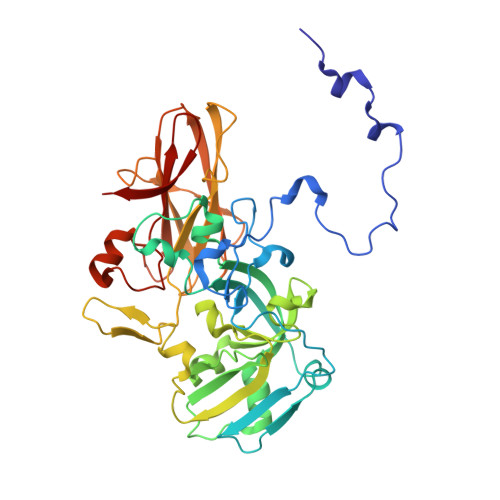
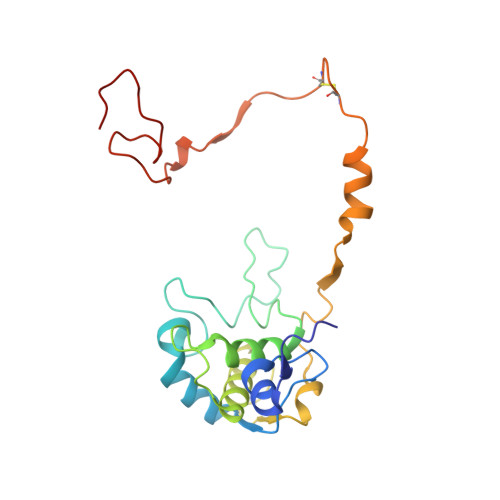
Structural Basis for the Oxidation of Protein-Bound Sulfur by the Sulfur Cycle Molybdohemo-Enzyme Sulfane Dehydrogenase Soxcd.
Zander, U., Faust, A., Klink, B.U., De Sanctis, D., Panjikar, S., Quentmeier, A., Bardischewsky, F., Friedrich, C.G., Scheidig, A.J.(2011) J Biological Chem 286: 8349
- PubMed: 21147779
- DOI: https://doi.org/10.1074/jbc.M110.193631
- Primary Citation of Related Structures:
2XTS - PubMed Abstract:
The sulfur cycle enzyme sulfane dehydrogenase SoxCD is an essential component of the sulfur oxidation (Sox) enzyme system of Paracoccus pantotrophus. SoxCD catalyzes a six-electron oxidation reaction within the Sox cycle. SoxCD is an α(2)β(2) heterotetrameric complex of the molybdenum cofactor-containing SoxC protein and the diheme c-type cytochrome SoxD with the heme domains D(1) and D(2). SoxCD(1) misses the heme-2 domain D(2) and is catalytically as active as SoxCD. The crystal structure of SoxCD(1) was solved at 1.33 Å. The substrate of SoxCD is the outer (sulfane) sulfur of Cys-110-persulfide located at the C-terminal peptide swinging arm of SoxY of the SoxYZ carrier complex. The SoxCD(1) substrate funnel toward the molybdopterin is narrow and partially shielded by side-chain residues of SoxD(1). For access of the sulfane-sulfur of SoxY-Cys-110 persulfide we propose that (i) the blockage by SoxD-Arg-98 is opened via interaction with the C terminus of SoxY and (ii) the C-terminal peptide VTIGGCGG of SoxY provides interactions with the entrance path such that the cysteine-bound persulfide is optimally positioned near the molybdenum atom. The subsequent oxidation reactions of the sulfane-sulfur are initiated by the nucleophilic attack of the persulfide anion on the molybdenum atom that is, in turn, reduced. The close proximity of heme-1 to the molybdopterin allows easy acceptance of the electrons. Because SoxYZ, SoxXA, and SoxB are already structurally characterized, with SoxCD(1) the structures of all key enzymes of the Sox cycle are known with atomic resolution.
- From the Department of Structural Biology, Zoological Institute, Christian-Albrechts-University Kiel, 24118 Kiel, Germany,; the Department of Biophysics-Structural Biology, Saarland University, 66421 Homburg, Germany.
Organizational Affiliation: