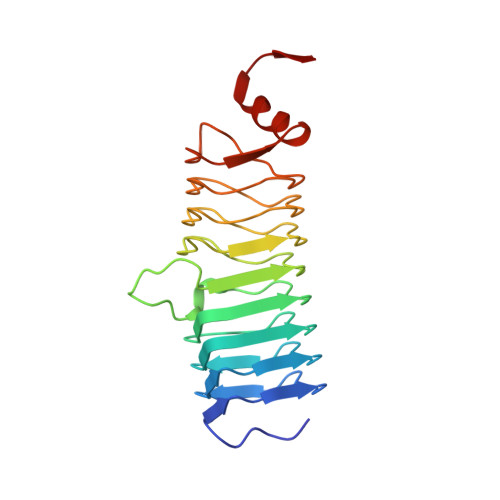
Pentapeptide-Repeat Proteins that Act as Topoisomerase Poison Resistance Factors Have a Common Dimer Interface.
Vetting, M.W., Hegde, S.S., Zhang, Y., Blanchard, J.S.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 296
- PubMed: 21393830
- DOI: https://doi.org/10.1107/S1744309110053315
- Primary Citation of Related Structures:
2XT2, 2XT4 - PubMed Abstract:
The protein AlbG is a self-resistance factor against albicidin, a nonribosomally encoded hybrid polyketide-peptide with antibiotic and phytotoxic properties produced by Xanthomonas albilineans. Primary-sequence analysis indicates that AlbG is a member of the pentapeptide-repeat family of proteins (PRP). The structure of AlbG from X. albilineans was determined at 2.0 Å resolution by SAD phasing using data collected from a single trimethyllead acetate derivative on a home source. AlbG folds into a right-handed quadrilateral β-helix composed of approximately eight semi-regular coils. The regularity of the β-helix is blemished by a large loop/deviation in the β-helix between coils 4 and 5. The C-terminus of the β-helix is capped by a dimerization module, yielding a dimer with a 110 Å semi-collinear β-helical axis. This method of dimer formation appears to be common to all PRP proteins that confer resistance to topoisomerase poisons and contrasts with most PRP proteins, which are typically monomeric.
- Department of Biochemistry, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461, USA. vetting@aecom.yu.edu
Organizational Affiliation: