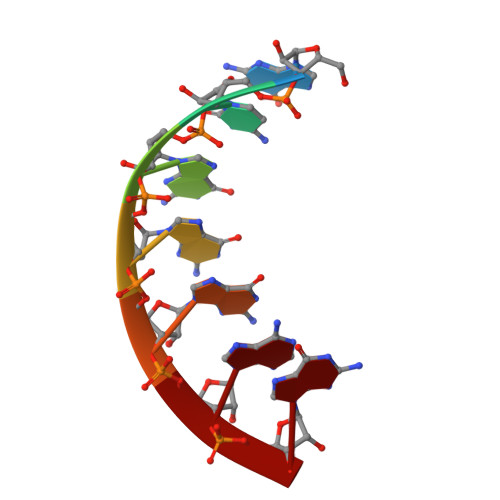
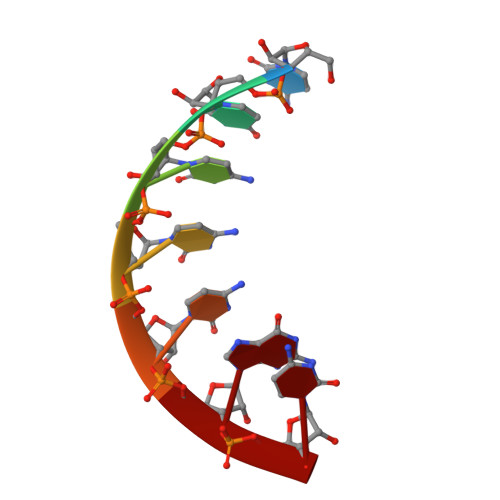
The Crystal Structure of a Thermus Thermophilus tRNA(Gly) Acceptor Stem Microhelix at 1.6 A Resolution.
Oberthur, D., Eichert, A., Erdmann, V.A., Furste, J.P., Betzel, C., Forster, C.(2011) Biochem Biophys Res Commun 404: 245
- PubMed: 21114959
- DOI: https://doi.org/10.1016/j.bbrc.2010.11.101
- Primary Citation of Related Structures:
2XSL - PubMed Abstract:
tRNAs are aminoacylated with the correct amino acid by the cognate aminoacyl-tRNA synthetase. The tRNA/synthetase systems can be divided into two classes: class I and class II. Within class I, the tRNA identity elements that enable the specificity consist of complex sequence and structure motifs, whereas in class II the identity elements are assured by few and simple determinants, which are mostly located in the tRNA acceptor stem. The tRNA(Gly)/glycyl-tRNA-synthetase (GlyRS) system is a special case regarding evolutionary aspects. There exist two different types of GlyRS, namely an archaebacterial/human type and an eubacterial type, reflecting the evolutionary divergence within this system. We previously reported the crystal structures of an Escherichia coli and of a human tRNA(Gly) acceptor stem microhelix. Here we present the crystal structure of a thermophilic tRNA(Gly) aminoacyl stem from Thermus thermophilus at 1.6Å resolution and provide insight into the RNA geometry and hydration.
- Institute of Biochemistry and Molecular Biology, Laboratory for Structural Biology of Infection and Inflammation, University of Hamburg, c/o DESY, 22603 Hamburg, Germany.
Organizational Affiliation: