Engineering a High Affinity Anti-Il-15 Antibody: Crystal Structure Reveals an Alpha-Helix in Vh Cdr3 as Key Component of Paratope.
Lowe, D.C., Gerhardt, S., Ward, A., Hargreaves, D., Anderson, M., Ferraro, F., Pauptit, R.A., Pattison, D.V., Buchanan, C., Popovic, B., Finch, D.K., Wilkinson, T., Sleeman, M., Vaughan, T.J., Mallinder, P.R.(2011) J Mol Biology 406: 160
- PubMed: 21167836
- DOI: https://doi.org/10.1016/j.jmb.2010.12.017
- Primary Citation of Related Structures:
2XQB - PubMed Abstract:
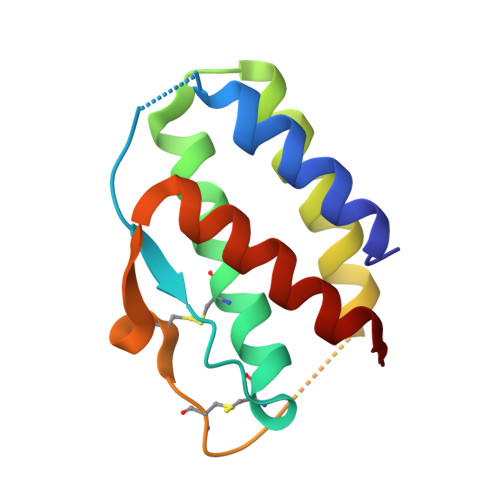
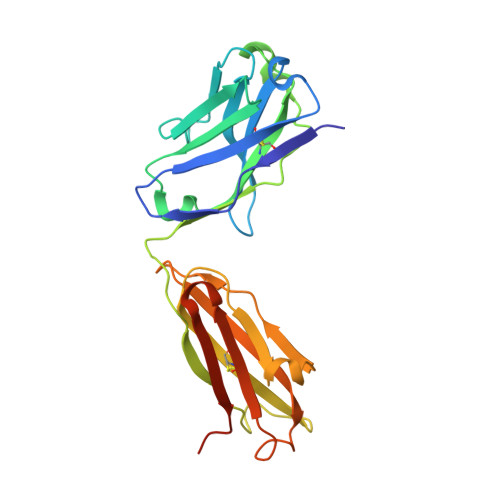
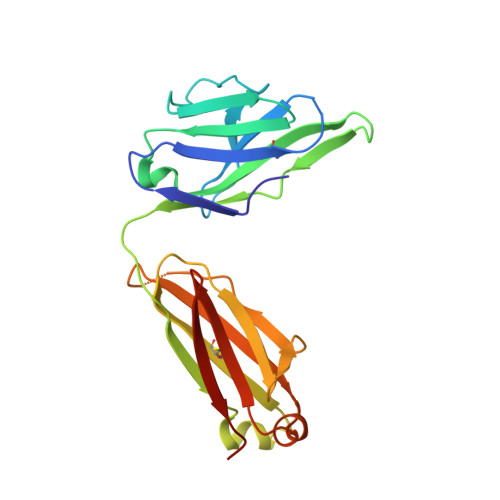
Interleukin (IL) 15 is an inflammatory cytokine that plays an essential role in the activation, proliferation, and maintenance of specific natural killer cell and T-cell populations, and has been implicated as a mediator of inflammatory diseases. An anti-IL-15 antibody that blocked IL-15-dependent cellular responses was isolated by phage display and optimised via mutagenesis of the third complementarity-determining regions (CDRs) of variable heavy (VH) and variable light chains. Entire repertoires of improved variants were recombined with each other to explore the maximum potential sequence space. DISC0280, the most potent antibody isolated using this comprehensive strategy, exhibits a 228-fold increase in affinity and a striking 40,000-fold increase in cellular potency compared to its parent. Such a wholesale recombination strategy therefore represents a useful method for exploiting synergistic potency gains as part of future antibody engineering efforts. The crystal structure of DISC0280 Fab (fragment antigen binding), in complex with human IL-15, was determined in order to map the structural epitope and paratope. The most remarkable feature revealed lies within the paratope and is a novel six-amino-acid α-helix that sits within the VH CDR3 loop at the center of the antigen binding site. This is the first report to describe an α-helix as a principal component of a naturally derived VH CDR3 following affinity maturation.
- MedImmune Ltd., Milstein Building, Granta Park, Cambridge CB1 6GH, UK. lowed@medimmune.com
Organizational Affiliation: