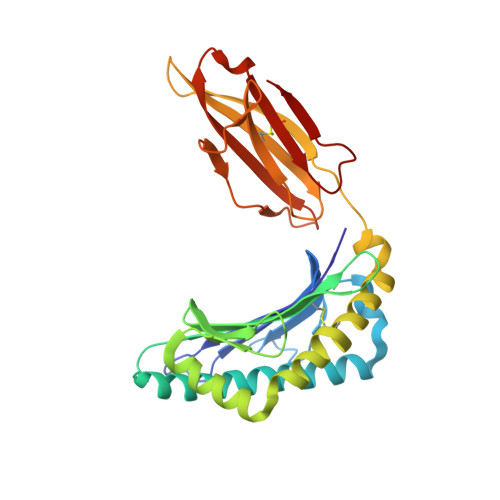
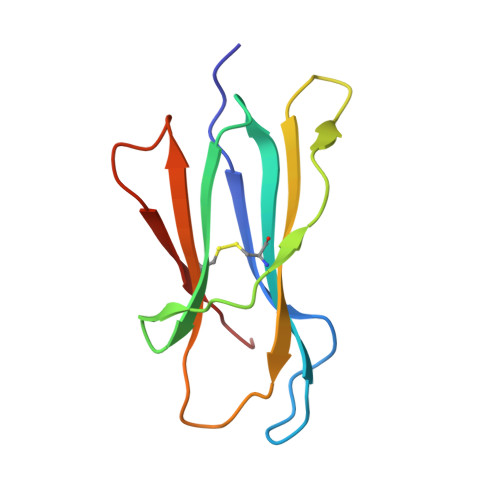
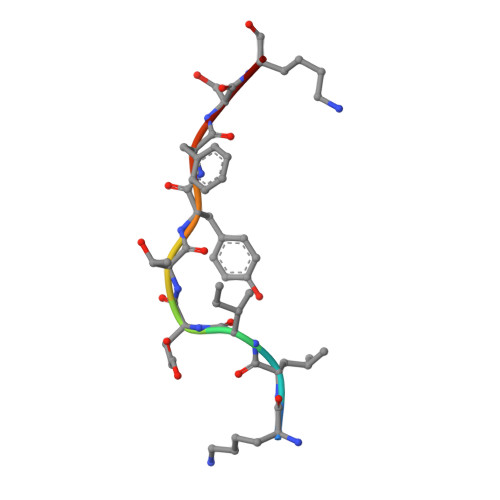
Structure of Hla-A0301 in Complex with a Peptide of Proteolipid Protein: Insights Into the Role of Hla-A Alleles in Susceptibility to Multiple Sclerosis
Mcmahon, R.M., Friis, L., Siebold, C., Friese, M.A., Fugger, L., Jones, E.Y.(2011) Acta Crystallogr D Biol Crystallogr 67: 447
- PubMed: 21543847
- DOI: https://doi.org/10.1107/S0907444911007888
- Primary Citation of Related Structures:
2XPG - PubMed Abstract:
The structure of the human major histocompatability (MHC) class I molecule HLA-A*0301 (HLA-A3) in complex with a nonameric peptide (KLIETYFSK) has been determined by X-ray crystallography to 2.7 Å resolution. HLA-A3 is a predisposing allele for multiple sclerosis (MS), an autoimmune disease of the central nervous system. The KLIETYFSK peptide is a naturally processed epitope of proteolipid protein, a myelin protein and candidate target for immune-mediated myelin destruction in MS. Comparison of the structure of HLA-A3 with that of HLA-A2, an MHC class I molecule which is protective against MS, indicates that both MHC class I molecules present very similar faces for T-cell receptor recognition whilst differing in the specificity of their peptide-binding grooves. These characteristics may underlie the opposing (predisposing versus protective) associations that they exhibit both in humans and in mouse models of MS-like disease. Furthermore, subtle alterations within the peptide-binding groove of HLA-A3 and other A3-like MHC class I molecules, members of the so-called A3 superfamily, may be sufficient to alter their presentation of autoantigen peptides such as KLIETYFSK. This in turn may modulate their contribution to the associated risk of autoimmune disease.
- Medical Research Council Human Immunology Unit, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, University of Oxford, Oxford OX39DS, England.
Organizational Affiliation: