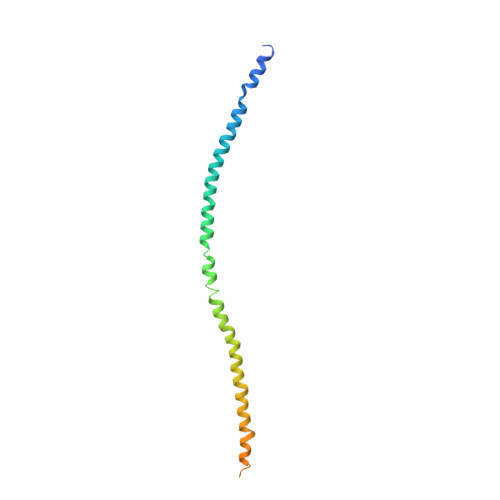Streptococcal M1 Protein Constructs a Pathological Host Fibrinogen Network
Macheboeuf, P., Buffalo, C., Fu, C.Y., Zinkernagel, A.S., Cole, J.N., Johnson, J.E., Nizet, V., Nizet, V., Ghosh, P.(2011) Nature 472: 64
- PubMed: 21475196
- DOI: https://doi.org/10.1038/nature09967
- Primary Citation of Related Structures:
2XNX, 2XNY - PubMed Abstract:
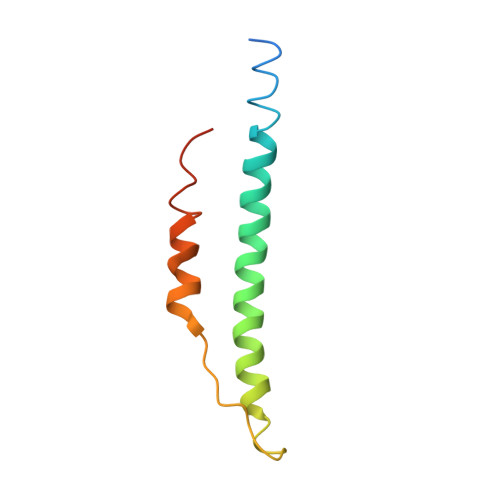
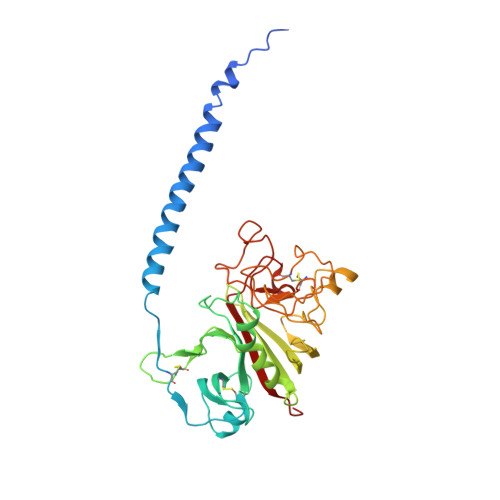
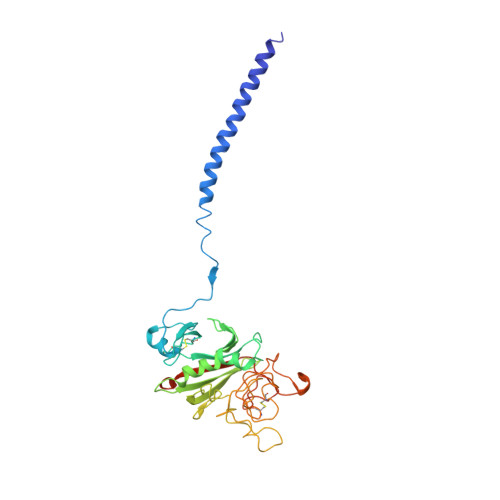
M1 protein, a major virulence factor of the leading invasive strain of group A Streptococcus, is sufficient to induce toxic-shock-like vascular leakage and tissue injury. These events are triggered by the formation of a complex between M1 and fibrinogen that, unlike M1 or fibrinogen alone, leads to neutrophil activation. Here we provide a structural explanation for the pathological properties of the complex formed between streptococcal M1 and human fibrinogen. A conformationally dynamic coiled-coil dimer of M1 was found to organize four fibrinogen molecules into a specific cross-like pattern. This pattern supported the construction of a supramolecular network that was required for neutrophil activation but was distinct from a fibrin clot. Disruption of this network into other supramolecular assemblies was not tolerated. These results have bearing on the pathophysiology of streptococcal toxic shock.
- Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, California 92093, USA.
Organizational Affiliation: