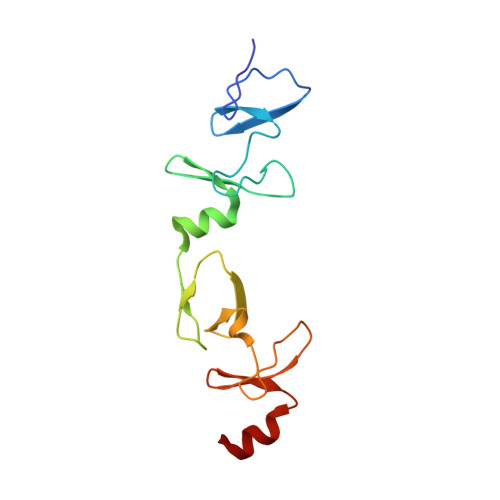
Structure of the Leukemia Oncogene Lmo2: Implications for the Assembly of a Hematopoietic Transcription Factor Complex.
El Omari, K., Hoosdally, S.J., Tuladhar, K., Karia, D., Vyas, P., Patient, R., Porcher, C., Mancini, E.J.(2011) Blood 117: 2146
- PubMed: 21076045
- DOI: https://doi.org/10.1182/blood-2010-07-293357
- Primary Citation of Related Structures:
2XJY, 2XJZ - PubMed Abstract:
The LIM only protein 2 (LMO2) is a key regulator of hematopoietic stem cell development whose ectopic expression in T cells leads to the onset of acute lymphoblastic leukemia. Through its LIM domains, LMO2 is thought to function as the scaffold for a DNA-binding transcription regulator complex, including the basic helix-loop-helix proteins SCL/TAL1 and E47, the zinc finger protein GATA-1, and LIM-domain interacting protein LDB1. To understand the role of LMO2 in the formation of this complex and ultimately to dissect its function in normal and aberrant hematopoiesis, we solved the crystal structure of LMO2 in complex with the LID domain of LDB1 at 2.4 Å resolution. We observe a largely unstructured LMO2 kept in register by the LID binding both LIM domains. Comparison of independently determined crystal structures of LMO2 reveals large movements around a conserved hinge between the LIM domains. We demonstrate that such conformational flexibility is necessary for binding of LMO2 to its partner protein SCL/TAL1 in vitro and for the function of this complex in vivo. These results, together with molecular docking and analysis of evolutionarily conserved residues, yield the first structural model of the DNA-binding complex containing LMO2, LDB1, SCL/TAL1, and GATA-1.
- Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK.
Organizational Affiliation: