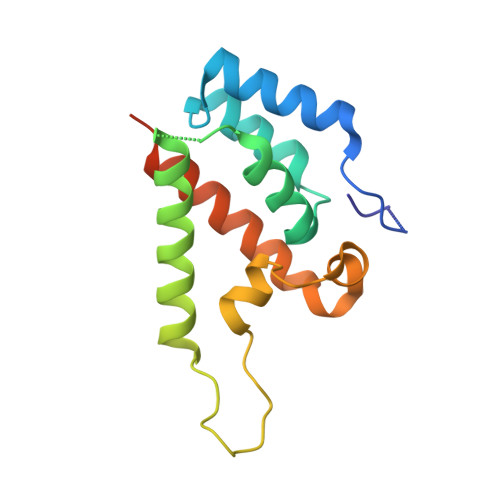
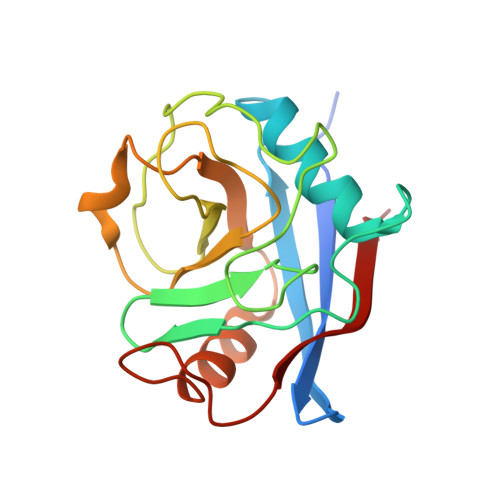
Structural and Functional Analysis of Prehistoric Lentiviruses Uncovers an Ancient Molecular Interface.
Goldstone, D.C., Yap, M.W., Robertson, L.E., Haire, L.F., Taylor, W.R., Katzourakis, A., Stoye, J.P., Taylor, I.A.(2010) Cell Host Microbe 8: 248
- PubMed: 20833376
- DOI: https://doi.org/10.1016/j.chom.2010.08.006
- Primary Citation of Related Structures:
2XGU, 2XGV, 2XGY - PubMed Abstract:
Lentiviruses are widespread in a variety of vertebrates, often associated with chronic disease states. However, until the recent discovery of the prehistoric endogenous lentiviruses in rabbits (RELIK) and lemurs (PSIV), it was thought that lentiviruses had no capacity for germline integration and were only spread horizontally in an exogenous fashion. The existence of RELIK and PSIV refuted these ideas, revealing lentiviruses to be present in a range of mammals, capable of germline integration, and far more ancient than previously thought. Using Gag sequences reconstructed from the remnants of these prehistoric lentiviruses, we have produced chimeric lentiviruses capable of infecting nondividing cells and determined structures of capsid domains from PSIV and RELIK. We show that the structures from these diverse viruses are highly similar, containing features found in modern-day lentiviruses, including a functional cyclophilin-binding loop. Together, these data provide evidence for an ancient capsid-cyclophilin interaction preserved throughout lentiviral evolution.
- Division of Molecular Structure, MRC National Institute for Medical Research, The Ridgeway, Mill Hill, London, UK.
Organizational Affiliation: