Mhc Class I Bound to an Immunodominant Theileria Parva Epitope Demonstrates Unconventional Presentation to T Cell Receptors.
Macdonald, I.K., Harkiolaki, M., Hunt, L., Connelley, T., Carroll, A.V., Machugh, N.D., Graham, S.P., Jones, E.Y., Morrison, W.I., Flower, D.R., Ellis, S.A.(2010) PLoS Pathog 6: 01149
- PubMed: 20976198
- DOI: https://doi.org/10.1371/journal.ppat.1001149
- Primary Citation of Related Structures:
2XFX - PubMed Abstract:
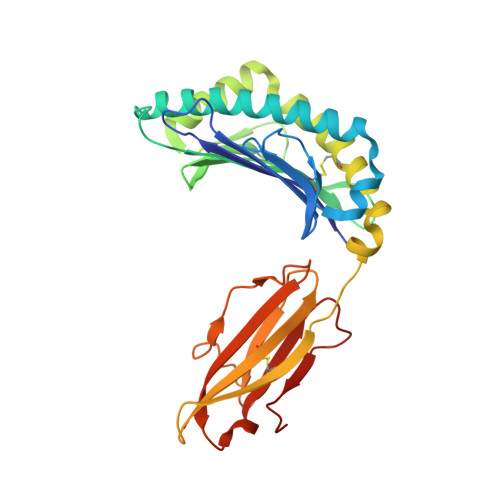
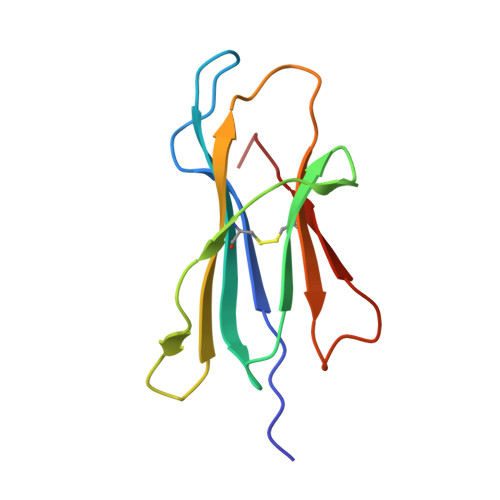
T cell receptor (TCR) recognition of peptide-MHC class I (pMHC) complexes is a crucial event in the adaptive immune response to pathogens. Peptide epitopes often display a strong dominance hierarchy, resulting in focusing of the response on a limited number of the most dominant epitopes. Such T cell responses may be additionally restricted by particular MHC alleles in preference to others. We have studied this poorly understood phenomenon using Theileria parva, a protozoan parasite that causes an often fatal lymphoproliferative disease in cattle. Despite its antigenic complexity, CD8+ T cell responses induced by infection with the parasite show profound immunodominance, as exemplified by the Tp1(214-224) epitope presented by the common and functionally important MHC class I allele N*01301. We present a high-resolution crystal structure of this pMHC complex, demonstrating that the peptide is presented in a distinctive raised conformation. Functional studies using CD8+ T cell clones show that this impacts significantly on TCR recognition. The unconventional structure is generated by a hydrophobic ridge within the MHC peptide binding groove, found in a set of cattle MHC alleles. Extremely rare in all other species, this feature is seen in a small group of mouse MHC class I molecules. The data generated in this analysis contribute to our understanding of the structural basis for T cell-dependent immune responses, providing insight into what determines a highly immunogenic p-MHC complex, and hence can be of value in prediction of antigenic epitopes and vaccine design.
- The Jenner Institute, University of Oxford, Compton, Berkshire, United Kingdom.
Organizational Affiliation: