Structural and Functional Characterization of Salmonella Enterica Serovar Typhimurium Ycbl: An Unusual Type II Glyoxalase
Stamp, A., Owen, P., El Omari, K., Nichols, C., Lockyer, M., Lamb, H., Charles, I., Hawkins, A.R., Stammers, D.K.(2010) Protein Sci 19: 1897
- PubMed: 20669241
- DOI: https://doi.org/10.1002/pro.475
- Primary Citation of Related Structures:
2XF4 - PubMed Abstract:
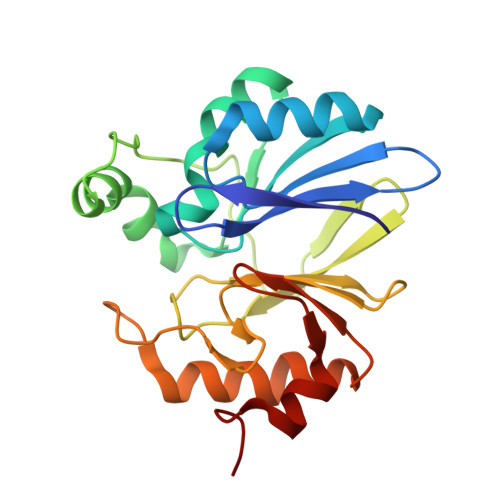
YcbL has been annotated as either a metallo-β-lactamase or glyoxalase II (GLX2), both members of the zinc metallohydrolase superfamily, that contains many enzymes with a diverse range of activities. Here, we report crystallographic and biochemical data for Salmonella enterica serovar Typhimurium YcbL that establishes it as GLX2, which differs in certain structural and functional properties compared with previously known examples. These features include the insertion of an α-helix after residue 87 in YcbL and truncation of the C-terminal domain, which leads to the loss of some recognition determinants for the glutathione substrate. Despite these changes, YcbL has robust GLX2 activity. A further difference is that the YcbL structure contains only a single bound metal ion rather than the dual site normally observed for GLX2s. Activity assays in the presence of various metal ions indicate an increase in activity above basal levels in the presence of manganous and ferrous ions. Thus, YcbL represents a novel member of the GLX2 family.
- Division of Structural Biology, The Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, United Kingdom.
Organizational Affiliation: