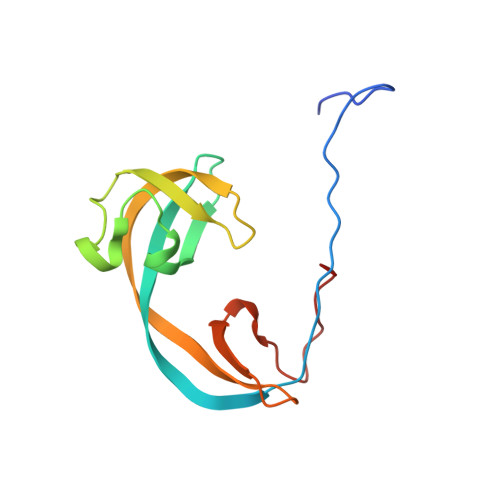
Crystal Structure of the Tudor Domain of Human Jmjd2C
Yue, W.W., Gileadi, C., Krojer, T., Weisbach, H., Ugochukwu, E., Daniel, M., Phillips, C., Chaikuad, A., von Delft, F., Allerston, C., Arrowsmith, C., Weigelt, J., Edwards, A., Bountra, C., Oppermann, U.To be published.