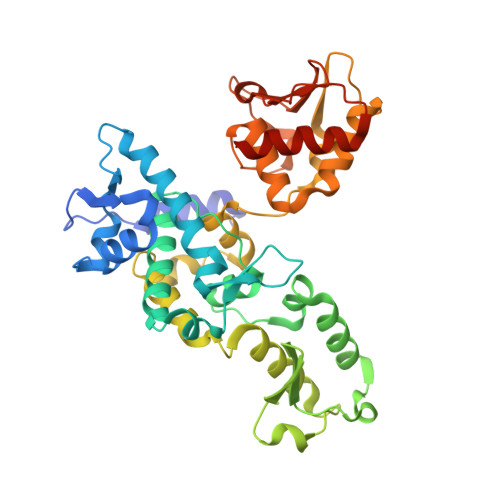
Structure of the Hect:Ubiquitin Complex and its Role in Ubiquitin Chain Elongation
Maspero, E., Mari, S., Valentini, E., Musacchio, A., Fish, A., Pasqualato, S., Polo, S.(2011) EMBO Rep 12: 342
- PubMed: 21399620
- DOI: https://doi.org/10.1038/embor.2011.21
- Primary Citation of Related Structures:
2XBB, 2XBF - PubMed Abstract:
Several mechanisms have been proposed for the synthesis of substrate-linked ubiquitin chains. HECT ligases directly catalyse protein ubiquitination and have been found to non-covalently interact with ubiquitin. We report crystal structures of the Nedd4 HECT domain, alone and in complex with ubiquitin, which show a new binding mode involving two surfaces on ubiquitin and both subdomains of the HECT N-lobe. The structures suggest a model for HECT-to-substrate ubiquitin transfer, in which the growing chain on the substrate is kept close to the catalytic cysteine to promote processivity. Mutational analysis highlights differences between the processes of substrate polyubiquitination and self-ubiquitination.
- IFOM, Fondazione Istituto FIRC di Oncologia Molecolare, Istituto Europeo di Oncologia, Via Adamello 16, Milan 20139, Italy.
Organizational Affiliation: