Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
Binda, C., Valente, S., Romanenghi, M., Pilotto, S., Cirilli, R., Karytinos, A., Ciossani, G., Botrugno, O.A., Forneris, F., Tardugno, M., Edmondson, D.E., Minucci, S., Mattevi, A., Mai, A.(2010) J Am Chem Soc 132: 6827
- PubMed: 20415477
- DOI: https://doi.org/10.1021/ja101557k
- Primary Citation of Related Structures:
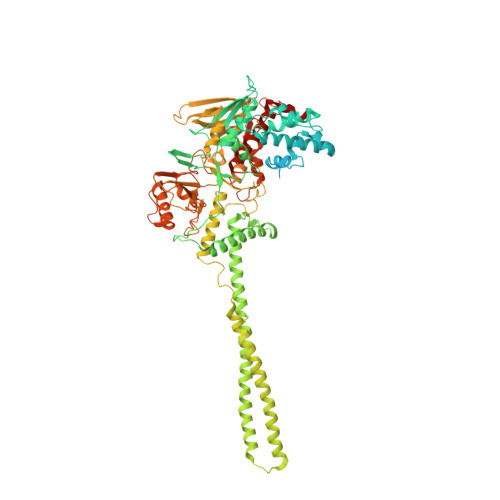
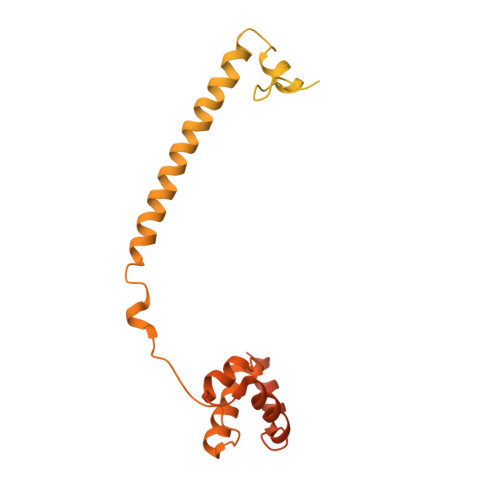
2XAF, 2XAG, 2XAH, 2XAJ, 2XAQ, 2XAS - PubMed Abstract:
LSD1 and LSD2 histone demethylases are implicated in a number of physiological and pathological processes, ranging from tumorigenesis to herpes virus infection. A comprehensive structural, biochemical, and cellular study is presented here to probe the potential of these enzymes for epigenetic therapies. This approach employs tranylcypromine as a chemical scaffold for the design of novel demethylase inhibitors. This drug is a clinically validated antidepressant known to target monoamine oxidases A and B. These two flavoenzymes are structurally related to LSD1 and LSD2. Mechanistic and crystallographic studies of tranylcypromine inhibition reveal a lack of selectivity and differing covalent modifications of the FAD cofactor depending on the enantiomeric form. These findings are pharmacologically relevant, since tranylcypromine is currently administered as a racemic mixture. A large set of tranylcypromine analogues were synthesized and screened for inhibitory activities. We found that the common evolutionary origin of LSD and MAO enzymes, despite their unrelated functions and substrate specificities, is reflected in related ligand-binding properties. A few compounds with partial enzyme selectivity were identified. The biological activity of one of these new inhibitors was evaluated with a cellular model of acute promyelocytic leukemia chosen since its pathogenesis includes aberrant activities of several chromatin modifiers. Marked effects on cell differentiation and an unprecedented synergistic activity with antileukemia drugs were observed. These data demonstrate that these LSD1/2 inhibitors are of potential relevance for the treatment of promyelocytic leukemia and, more generally, as tools to alter chromatin state with promise of a block of tumor progression.
- Department of Genetics and Microbiology, University of Pavia, Via Ferrata 1, 27100 Pavia, Italy.
Organizational Affiliation: