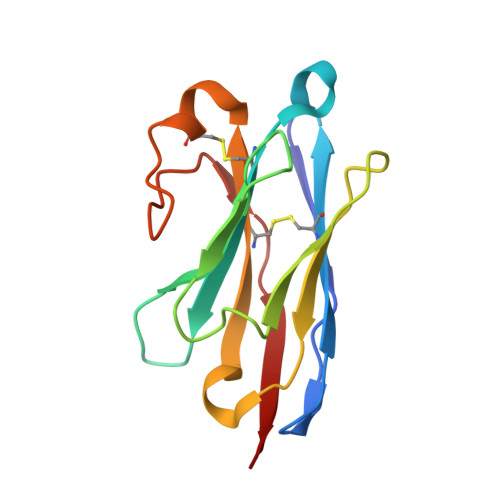
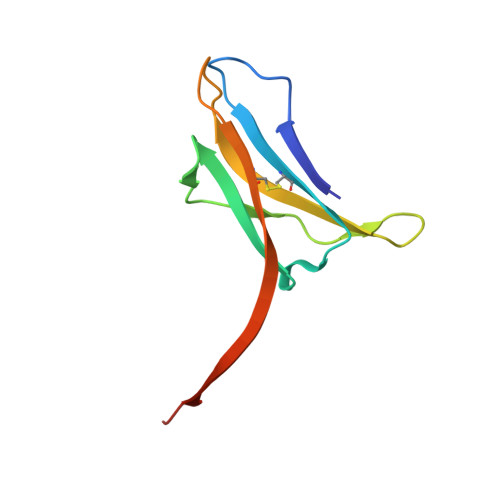
Atomic Structure of a Nanobody-Trapped Domain-Swapped Dimer of an Amyloidogenic {Beta}2-Microglobulin Variant.
Domanska, K., Vanderhaegen, S., Srinivasan, V., Pardon, E., Dupeux, F., Marquez, J.A., Giorgetti, S., Stoppini, M., Wyns, L., Bellotti, V., Steyaert, J.(2011) Proc Natl Acad Sci U S A 108: 1314
- PubMed: 21220305
- DOI: https://doi.org/10.1073/pnas.1008560108
- Primary Citation of Related Structures:
2X89 - PubMed Abstract:
Atomic-level structural investigation of the key conformational intermediates of amyloidogenesis remains a challenge. Here we demonstrate the utility of nanobodies to trap and characterize intermediates of β2-microglobulin (β2m) amyloidogenesis by X-ray crystallography. For this purpose, we selected five single domain antibodies that block the fibrillogenesis of a proteolytic amyloidogenic fragment of β2m (ΔN6β2m). The crystal structure of ΔN6β2m in complex with one of these nanobodies (Nb24) identifies domain swapping as a plausible mechanism of self-association of this amyloidogenic protein. In the swapped dimer, two extended hinge loops--corresponding to the heptapetide NHVTLSQ that forms amyloid in isolation--are unmasked and fold into a new two-stranded antiparallel β-sheet. The β-strands of this sheet are prone to self-associate and stack perpendicular to the direction of the strands to build large intermolecular β-sheets that run parallel to the axis of growing oligomers, providing an elongation mechanism by self-templated growth.
- Department of Molecular and Cellular Interactions, Vlaams Instituut voor Biotechnologie, Pleinlaan 2, B-1050 Brussels, Belgium.
Organizational Affiliation: