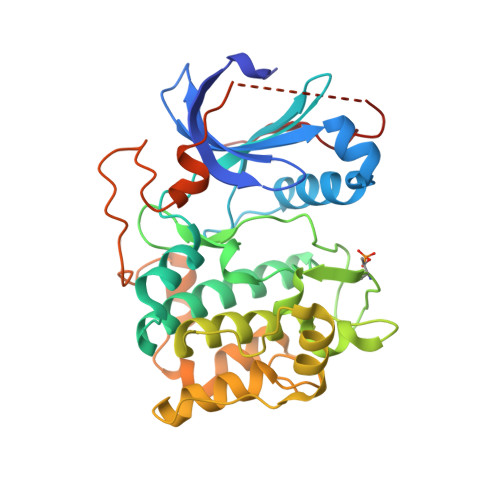
Discovery of 4-Amino-1-(7H-Pyrrolo[2,3-D]Pyrimidin-4-Yl)Piperidine-4-Carboxamides as Selective, Orally Active Inhibitors of Protein Kinase B (Akt).
Mchardy, T., Caldwell, J.J., Cheung, K.M., Hunter, L.J., Taylor, K., Rowlands, M., Ruddle, R., Henley, A., De Haven Brandon, A., Valenti, M., Davies, T.G., Fazal, L., Seavers, L., Raynaud, F.I., Eccles, S.A., Aherne, G.W., Garrett, M.D., Collins, I.(2010) J Med Chem 53: 2239
- PubMed: 20151677
- DOI: https://doi.org/10.1021/jm901788j
- Primary Citation of Related Structures:
2X39, 2XH5 - PubMed Abstract:
Protein kinase B (PKB or Akt) is an important component of intracellular signaling pathways regulating growth and survival. Signaling through PKB is frequently deregulated in cancer, and inhibitors of PKB therefore have potential as antitumor agents. The optimization of lipophilic substitution within a series of 4-benzyl-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-amines provided ATP-competitive, nanomolar inhibitors with up to 150-fold selectivity for inhibition of PKB over the closely related kinase PKA. Although active in cellular assays, compounds containing 4-amino-4-benzylpiperidines underwent metabolism in vivo, leading to rapid clearance and low oral bioavailability. Variation of the linker group between the piperidine and the lipophilic substituent identified 4-amino-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamides as potent and orally bioavailable inhibitors of PKB. Representative compounds modulated biomarkers of signaling through PKB in vivo and strongly inhibited the growth of human tumor xenografts in nude mice at well-tolerated doses.
- Cancer Research UK Centre for Cancer Therapeutics, The Institute of Cancer Research, Sutton, Surrey SM2 5NG, UK.
Organizational Affiliation: