Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
Najmudin, S., Pinheiro, B.A., Prates, J.A.M., Gilbert, H.J., Romao, M.J., Fontes, C.M.G.A.(2010) J Struct Biol 172: 353
- PubMed: 20682344
- DOI: https://doi.org/10.1016/j.jsb.2010.07.009
- Primary Citation of Related Structures:
2W5F, 2WYS, 2WZE - PubMed Abstract:
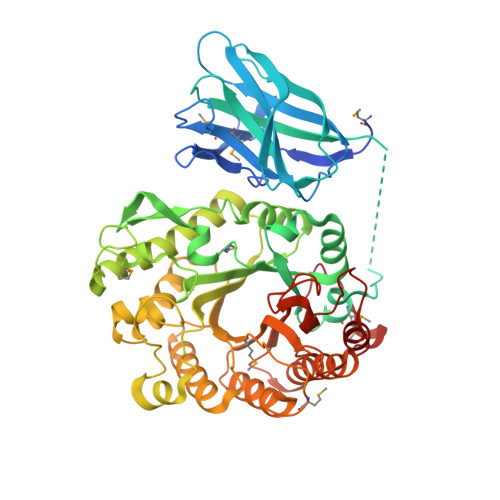
In general, plant cell wall degrading enzymes are modular proteins containing catalytic domains linked to one or more non-catalytic carbohydrate-binding modules (CBMs). Xyn10B from Clostridium thermocellum is a typical modular enzyme containing an N-terminal family 22 CBM (CBM22-1), a family 10 glycoside hydrolase catalytic domain (GH10), a second CBM22 (CBM22-2), a dockerin sequence and a C-terminal family 1 carbohydrate esterase (CE1) catalytic domain. The structure of the N-terminal bi-modular CBM22-1-GH10 component of Xyn10B has been determined using a SeMet derivative by SAD to 2.5Å. The data was extended to 2.0Å for the non-SeMet mutant complexed with xylohexaose. CBM22-1-GH10 is a 60kDa protein with an E337A mutation to render the GH10 subunit inactive. Three of the six xylose residues of xylohexaose are shown to be bound in the inactivated GH10 substrate binding cleft, with the other three sugars presumably disordered in the solvent channel. The protein is a dimer in the asymmetric unit with extensive surface contacts between the two GH10 modules and between the CBM22-1 and GH10 modules. Residues from helix H4 of the GH10 module provide the major contacts by fitting into the minor groove of the CBM22-1 module. The orientation of CBM22-1 is such that it would allow the substrate to be loosely bound and subsequently delivered to the active site in a processive manner.
- CIISA - Faculdade de Medicina Veterinária, Universidade Técnica de Lisboa, Avenida da Universidade Técnica, 1300-477 Lisboa, Portugal. shabir@fmv.utl.pt
Organizational Affiliation: