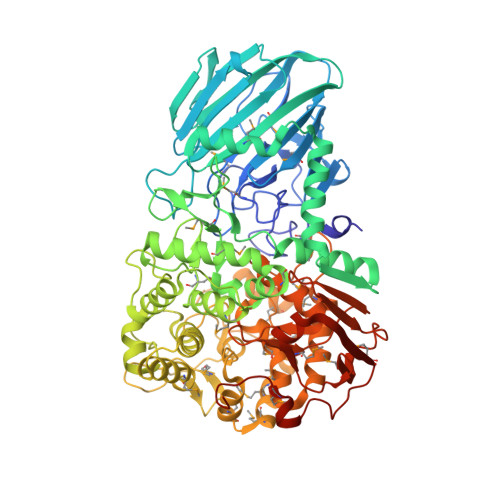
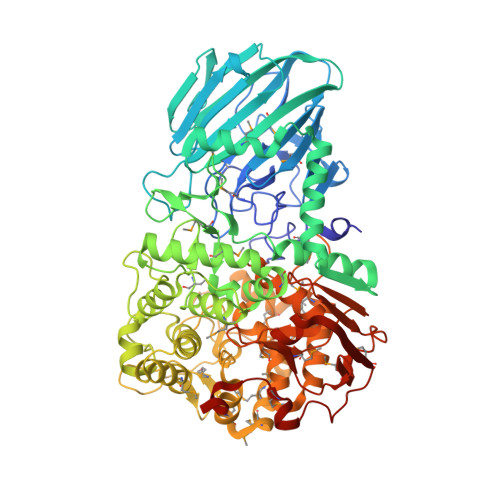
Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Zhu, Y., Suits, M.D.L., Thompson, A., Chavan, S., Dinev, Z., Dumon, C., Smith, N., Moremen, K.W., Xiang, Y., Siriwardena, A., Williams, S.J., Gilbert, H.J., Davies, G.J.(2010) Nat Chem Biol 6: 125
- PubMed: 20081828
- DOI: https://doi.org/10.1038/nchembio.278
- Primary Citation of Related Structures:
2WVX, 2WVY, 2WVZ, 2WW0, 2WW1, 2WW2, 2WW3, 2WZS - PubMed Abstract:
Colonic bacteria, exemplified by Bacteroides thetaiotaomicron, play a key role in maintaining human health by harnessing large families of glycoside hydrolases (GHs) to exploit dietary polysaccharides and host glycans as nutrients. Such GH family expansion is exemplified by the 23 family GH92 glycosidases encoded by the B. thetaiotaomicron genome. Here we show that these are alpha-mannosidases that act via a single displacement mechanism to utilize host N-glycans. The three-dimensional structure of two GH92 mannosidases defines a family of two-domain proteins in which the catalytic center is located at the domain interface, providing acid (glutamate) and base (aspartate) assistance to hydrolysis in a Ca(2+)-dependent manner. The three-dimensional structures of the GH92s in complex with inhibitors provide insight into the specificity, mechanism and conformational itinerary of catalysis. Ca(2+) plays a key catalytic role in helping distort the mannoside away from its ground-state (4)C(1) chair conformation toward the transition state.
- Institute for Cell and Molecular Biosciences, The Medical School, Newcastle University, Framlington Place, Newcastle upon Tyne, UK.
Organizational Affiliation: