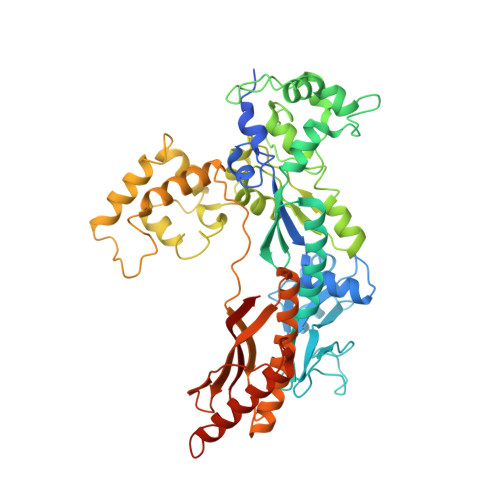
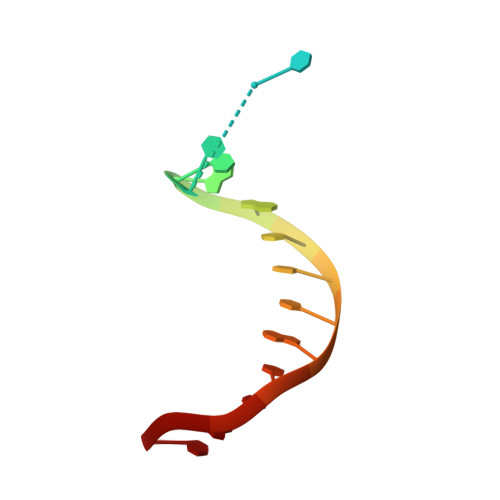
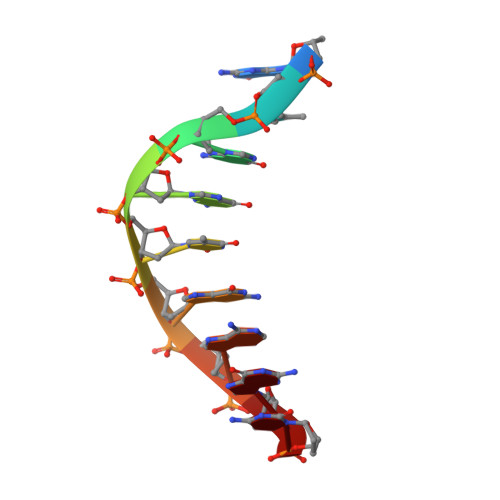
Crystal Structure of a Cisplatin-(1,3-Gtg) Cross-Link within DNA Polymerase Eta.
Reissner, T., Schneider, S., Schorr, S., Carell, T.(2010) Angew Chem Int Ed Engl 49: 3077
- PubMed: 20333640
- DOI: https://doi.org/10.1002/anie.201000414
- Primary Citation of Related Structures:
2WTF - Center for Integrated Protein Science (CiPSM), Department of Chemistry, Ludwig-Maximilians-University, Butenandtstrasse 5-13, 81377 Munich, Germany.
Organizational Affiliation: