Structural and Thermodynamic Characterization of the Adrenodoxin-Like Domain of the Electron-Transfer Protein Etp1 from Schizosaccharomyces Pombe.
Mueller, J.J., Hannemann, F., Schiffler, B., Ewen, K.M., Kappl, R., Heinemann, U., Bernhardt, R.(2011) J Inorg Biochem 105: 957
- PubMed: 21536008
- DOI: https://doi.org/10.1016/j.jinorgbio.2011.04.001
- Primary Citation of Related Structures:
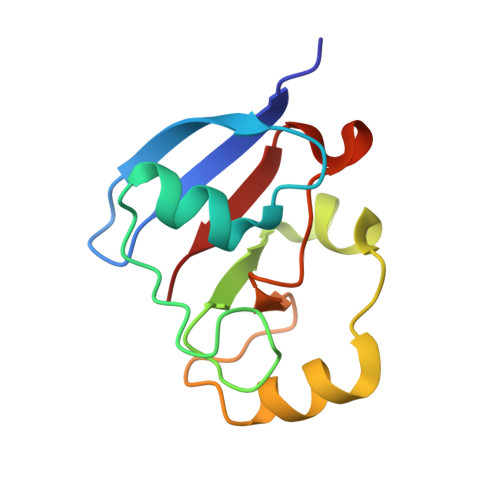
2WLB - PubMed Abstract:
The protein Etp1 of Schizosaccharomyces pombe consists of an amino-terminal COX15-like domain and a carboxy-terminal ferredoxin-like domain, Etp1(fd), which is cleaved off after mitochondrial import. The physiological function of Etp1(fd) is supposed to lie in the participation in the assembly of iron-sulfur clusters and the synthesis of heme A. In addition, the protein was shown to be the first microbial ferredoxin being able to support electron transfer in mitochondrial steroid hydroxylating cytochrome P450 systems in vivo and in vitro, replacing thereby the native redox partner, adrenodoxin. Despite a sequence similarity of 39% and the fact that fission yeast is a mesophilic organism, thermodynamic studies revealed that Etp1(fd) has a melting temperature more than 20°C higher than adrenodoxin. The three-dimensional structure of Etp1(fd) has been determined by crystallography. To the best of our knowledge it represents the first three-dimensional structure of a yeast ferredoxin. The structure-based sequence alignment of Etp1(fd) with adrenodoxin yields a rational explanation for their observed mutual exchangeability in the cytochrome P450 system. Analysis of the electron exchange with the S. pombe redox partner Arh1 revealed differences between Etp1(fd) and adrenodoxin, which might be linked to their different physiological functions in the mitochondria of mammals and yeast.
- Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany.
Organizational Affiliation: