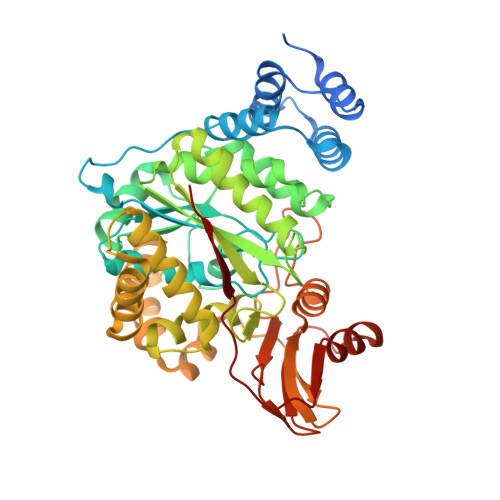
Structures of Native Human Thymidine Phosphorylase and in Complex with 5-Iodouracil.
Mitsiki, E., Papageorgiou, A.C., Iyer, S., Thiyagarajan, N., Prior, S.H., Sleep, D., Finnis, C., Acharya, K.R.(2009) Biochem Biophys Res Commun 386: 666
- PubMed: 19555658
- DOI: https://doi.org/10.1016/j.bbrc.2009.06.104
- Primary Citation of Related Structures:
2WK5, 2WK6 - PubMed Abstract:
Thymidine phosphorylase (TP) first identified as platelet derived endothelial cell growth factor (PD-ECGF) plays a key role in nucleoside metabolism. Human TP (hTP) is implicated in angiogenesis and is overexpressed in several solid tumors. Here, we report the crystal structures of recombinant hTP and its complex with a substrate 5-iodouracil (5IUR) at 3.0 and 2.5A, respectively. In addition, we provide information on the role of specific residues in the enzymatic activity of hTP through mutagenesis and kinetic studies.
- Department of Biology and Biochemistry, University of Bath, Claverton Down, Bath BA2 7AY, UK.
Organizational Affiliation: