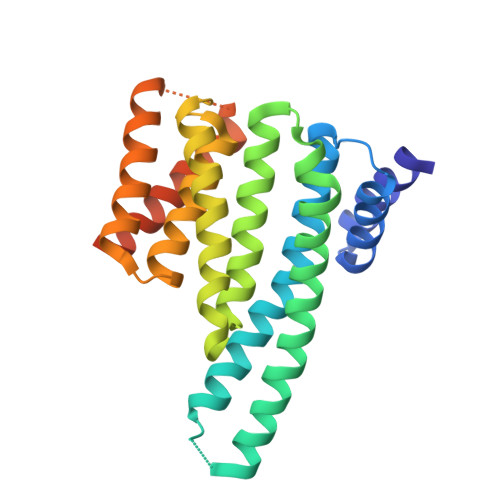
Recognition of an Intra-Chain Tandem 14-3-3 Binding Site within Pkc Epsilon.
Kostelecky, B., Saurin, A.T., Purkiss, A., Parker, P.J., Mcdonald, N.Q.(2009) EMBO Rep 10: 983
- PubMed: 19662078
- DOI: https://doi.org/10.1038/embor.2009.150
- Primary Citation of Related Structures:
2WH0 - PubMed Abstract:
The phosphoserine/threonine binding protein 14-3-3 stimulates the catalytic activity of protein kinase C-epsilon (PKCepsilon) by engaging two tandem phosphoserine-containing motifs located between the PKCepsilon regulatory and catalytic domains (V3 region). Interaction between 14-3-3 and this region of PKCepsilon is essential for the completion of cytokinesis. Here, we report the crystal structure of 14-3-3zeta bound to a synthetic diphosphorylated PKCepsilon V3 region revealing how a consensus 14-3-3 site and a divergent 14-3-3 site cooperate to bind to 14-3-3 and so activate PKCepsilon. Thermodynamic data show a markedly enhanced binding affinity for two-site phosphopeptides over single-site 14-3-3 binding motifs and identifies Ser 368 as a gatekeeper phosphorylation site in this physiologically relevant 14-3-3 ligand. This dual-site intra-chain recognition has implications for other 14-3-3 targets, which seem to have only a single 14-3-3 motif, as other lower affinity and cryptic 14-3-3 gatekeeper sites might exist.
- Structural Biology Laboratory, London Research Institute, Cancer Research UK, 44 Lincoln's Inn Fields, London WC2A 3PX, UK.
Organizational Affiliation: