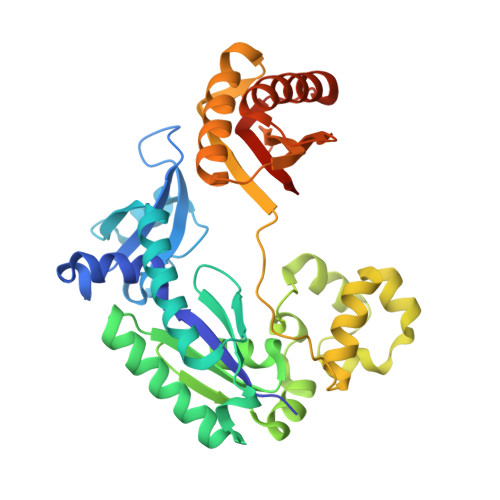
Structure-Function Relationships in Miscoding by Sulfolobus Solfataricus DNA Polymerase Dpo4: Guanine N2,N2-Dimethyl Substitution Produces Inactive and Miscoding Polymerase Complexes.
Zhang, H., Eoff, R.L., Kozekov, I.D., Rizzo, C.J., Egli, M., Guengerich, F.P.(2009) J Biological Chem 284: 17687
- PubMed: 19542237
- DOI: https://doi.org/10.1074/jbc.M109014274
- Primary Citation of Related Structures:
2W9A, 2W9B, 2W9C - PubMed Abstract:
Previous work has shown that Y-family DNA polymerases tolerate large DNA adducts, but a substantial decrease in catalytic efficiency and fidelity occurs during bypass of N2,N2-dimethyl (Me2)-substituted guanine (N2,N2-Me2G), in contrast to a single methyl substitution. Therefore, it is unclear why the addition of two methyl groups is so disruptive. The presence of N2,N2-Me2G lowered the catalytic efficiency of the model enzyme Sulfolobus solfataricus Dpo4 16,000-fold. Dpo4 inserted dNTPs almost at random during bypass of N2,N2-Me2G, and much of the enzyme was kinetically trapped by an inactive ternary complex when N2,N2-Me2G was present, as judged by a reduced burst amplitude (5% of total enzyme) and kinetic modeling. One crystal structure of Dpo4 with a primer having a 3'-terminal dideoxycytosine (Cdd) opposite template N2,N2-Me2G in a post-insertion position showed Cdd folded back into the minor groove, as a catalytically incompetent complex. A second crystal had two unique orientations for the primer terminal Cdd as follows: (i) flipped into the minor groove and (ii) a long pairing with N2,N2-Me2G in which one hydrogen bond exists between the O-2 atom of Cdd and the N-1 atom of N2,N2-Me2G, with a second water-mediated hydrogen bond between the N-3 atom of Cdd and the O-6 atom of N2,N2-Me2G. A crystal structure of Dpo4 with dTTP opposite template N2,N2-Me2G revealed a wobble orientation. Collectively, these results explain, in a detailed manner, the basis for the reduced efficiency and fidelity of Dpo4-catalyzed bypass of N2,N2-Me2G compared with mono-substituted N2-alkyl G adducts.
- Department of Biochemistry and Center in Molecular Toxicology, Vanderbilt University School of Medicine, Nashville, Tennessee 37232-0146, USA.
Organizational Affiliation: