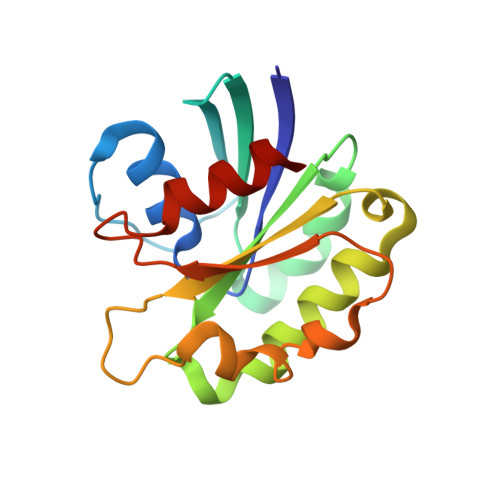
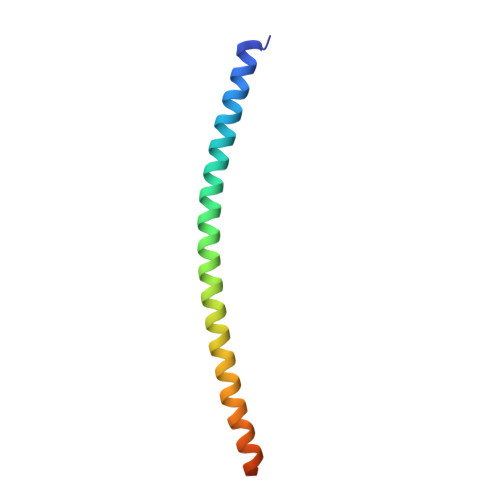The Structural Basis of Arf Effector Specificity: The Crystal Structure of Arf6 in a Complex with Jip4.
Isabet, T., Montagnac, G., Regazzoni, K., Raynal, B., El Khadali, F., England, P., Franco, M., Chavrier, P., Houdusse, A., Menetrey, J.(2009) EMBO J 28: 2835
- PubMed: 19644450
- DOI: https://doi.org/10.1038/emboj.2009.209
- Primary Citation of Related Structures:
2W83 - PubMed Abstract:
The JNK-interacting proteins, JIP3 and JIP4, are specific effectors of the small GTP-binding protein ARF6. The interaction of ARF6-GTP with the second leucine zipper (LZII) domains of JIP3/JIP4 regulates the binding of JIPs to kinesin-1 and dynactin. Here, we report the crystal structure of ARF6-GTP bound to the JIP4-LZII at 1.9 A resolution. The complex is a heterotetramer with dyad symmetry arranged in an ARF6-(JIP4)(2)-ARF6 configuration. Comparison of the ARF6-JIP4 interface with the equivalent region of ARF1 shows the structural basis of JIP4's specificity for ARF6. Using site-directed mutagenesis and surface plasmon resonance, we further show that non-conserved residues at the switch region borders are the key structural determinants of JIP4 specificity. A structure-derived model of the association of the ARF6-JIP3/JIP4 complex with membranes shows that the JIP4-LZII coiled-coil should lie along the membrane to prevent steric hindrances, resulting in only one ARF6 molecule bound. Such a heterotrimeric complex gives insights to better understand the ARF6-mediated motor switch regulatory function.
- Institut Curie, Centre de Recherche, Paris, France.
Organizational Affiliation: