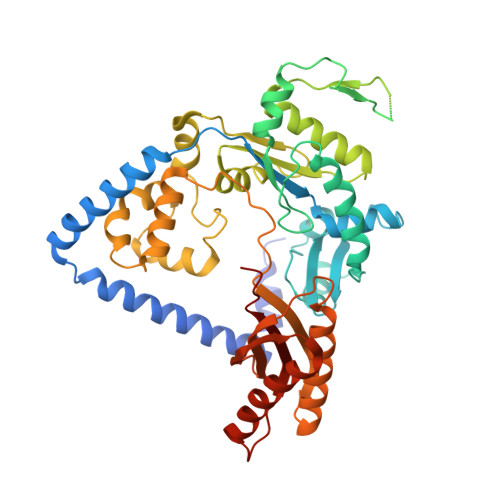
Structural and Functional Elucidation of the Mechanism Promoting Error-Prone Synthesis by Human DNA Polymerase Kappa Opposite the 7,8-Dihydro-8-Oxo-2'-Deoxyguanosine Adduct.
Irimia, A., Eoff, R.L., Guengerich, F.P., Egli, M.(2009) J Biological Chem 284: 22467
- PubMed: 19542228
- DOI: https://doi.org/10.1074/jbc.M109.003905
- Primary Citation of Related Structures:
2W7O, 2W7P - PubMed Abstract:
Human polymerase kappa (hPol kappa) is one of four eukaryotic Y-class DNA polymerases and may be an important element in the cellular response to polycyclic aromatic hydrocarbons such as benzo[a]pyrene, which can lead to reactive oxygenated metabolite-mediated oxidative stress. Here, we present a detailed analysis of the activity and specificity of hPol kappa bypass opposite the major oxidative adduct 7,8-dihydro-8-oxo-2'-deoxyguanosine (8-oxoG). Unlike its archaeal homolog Dpo4, hPol kappa bypasses this lesion in an error-prone fashion by inserting mainly dATP. Analysis of transient-state kinetics shows diminished "bursts" for dATP:8-oxoG and dCTP:8-oxoG incorporation, indicative of non-productive complex formation, but dATP:8-oxoG insertion events that do occur are 2-fold more efficient than dCTP:G insertion events. Crystal structures of ternary hPol kappa complexes with adducted template-primer DNA reveal non-productive (dGTP and dATP) alignments of incoming nucleotide and 8-oxoG. Structural limitations placed upon the hPol kappa by interactions between the N-clasp and finger domains combined with stabilization of the syn-oriented template 8-oxoG through the side chain of Met-135 both appear to contribute to error-prone bypass. Mutating Leu-508 in the little finger domain of hPol kappa to lysine modulates the insertion opposite 8-oxoG toward more accurate bypass, similar to previous findings with Dpo4. Our structural and activity data provide insight into important mechanistic aspects of error-prone bypass of 8-oxoG by hPol kappa compared with accurate and efficient bypass of the lesion by Dpo4 and polymerase eta.
- Department of Biochemistry and Center in Molecular Toxicology, Vanderbilt University School of Medicine, Nashville, Tennessee 37232-0146.
Organizational Affiliation: