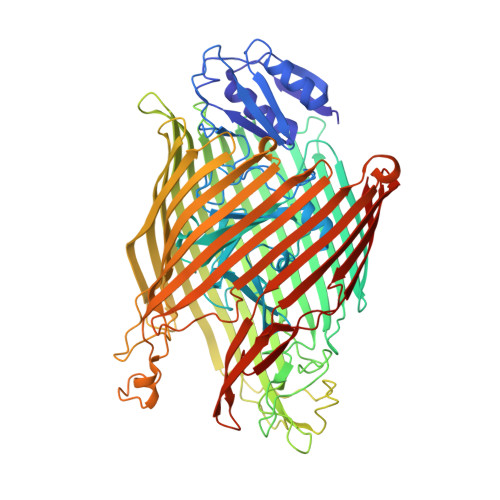
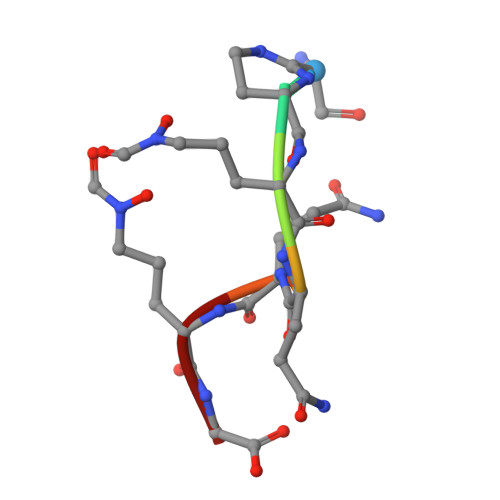
Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Greenwald, J., Nader, M., Celia, H., Gruffaz, C., Geoffroy, V., Meyer, J.M., Schalk, I.J., Pattus, F.(2009) Mol Microbiol 72: 1246
- PubMed: 19504741
- DOI: https://doi.org/10.1111/j.1365-2958.2009.06721.x
- Primary Citation of Related Structures:
2W16, 2W6T, 2W6U, 2W75, 2W76, 2W77, 2W78 - PubMed Abstract:
The first step in the specific uptake of iron via siderophores in Gram-negative bacteria is the recognition and binding of a ferric siderophore by its cognate receptor. We investigated the molecular basis of this event through structural and biochemical approaches. FpvA, the pyoverdine-Fe transporter from Pseudomonas aeruginosa ATCC 15692 (PAO1 strain), is able to transport ferric-pyoverdines originating from other species, whereas most fluorescent pseudomonads are only able to use the one they produce among the more than 100 known different pyoverdines. We solved the structure of FpvA bound to non-cognate pyoverdines of high- or low-affinity and found a close correlation between receptor-ligand structure and the measured affinities. The structure of the first amino acid residues of the pyoverdine chain distinguished the high- and low-affinity binders while the C-terminal portion of the pyoverdines, often cyclic, does not appear to contribute extensively to the interaction between the siderophore and its transporter. The specificity of the ferric-pyoverdine binding site of FpvA is conferred by the structural elements common to all ferric-pyoverdines, i.e. the chromophore, iron, and its chelating groups.
- Laboratoire de Biologie Structurale des Membranes, UMR7175, Ecole Supérieure de Biotechnologie de Strasbourg, Bd Sébastien Brant, BP10413, 67412 Illkirch, France. jason.greenwald@phys.chem.ethz.ch
Organizational Affiliation: