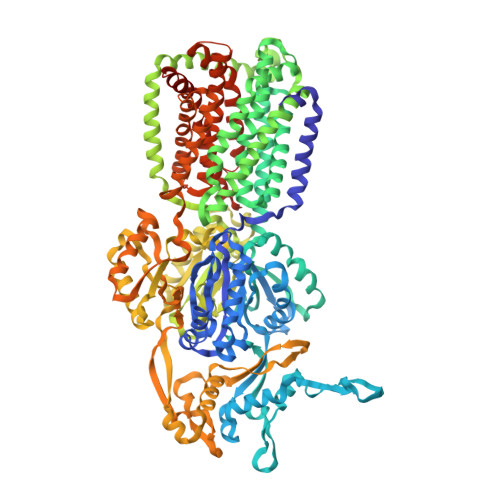
The Structure of the Efflux Pump Acrb in Complex with Bile Acid.
Drew, D., Klepsch, M.M., Newstead, S., Flaig, R., De Gier, J.W., Iwata, S., Beis, K.(2008) Mol Membr Biol 25: 677
- PubMed: 19023693
- DOI: https://doi.org/10.1080/09687680802552257
- Primary Citation of Related Structures:
2W1B - PubMed Abstract:
Gastrointestinal bacteria, like Escherichia coli, must remove bile acid to survive in the gut. Bile acid removal in E. coli is thought to be mediated primarily by the multidrug efflux pump, AcrB. Here, we present the structure of E. coli AcrB in complex with deoxycholate at 3.85 A resolution. All evidence suggests that bile acid is transported out of the cell via the periplasmic vestibule of the AcrAB-TolC complex.
- Division of Molecular Biosciences, Membrane Protein Crystallography Group and Membrane Protein Laboratory, Imperial College, London, UK.
Organizational Affiliation: